-
Bioclipse git experiences #1: Strip away unwanted plugins
This is a series of two posts repeating some content I wrote up back in the Bioclipse days (see also this Scholia page). They both deal with something we were facing: restructuring of version control repositories, while actually keeping the history. For example, you may want to copy or move code from one repository to another. A second use case can be a file that must be removed (there are valid reasons for that). Because these posts are based on Bioclipse work, there will be some specific terminology, but the approach I regularly apply in other situations. -
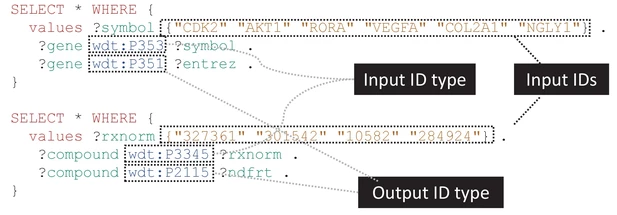
new paper: "Wikidata as a knowledge graph for the life sciences"
A figure from the article, outlining the idea of using SPARQL queries to extract data from the open knowledge base. As a reader of my blog, you know I have been doing quite some research where Wikidata has some role. I am preparing a paper on the work I have done around chemicals in Wikidata, based on what I presented at the ICCS with a poster. So, I was delighted when Andra and Andrew asked me to contribute to a paper outline the importance of Wikidata to the life sciences. The paper was published in eLife, which I’m excited about to, as they do a significant amount of publishing innovation. -

What metabolites are found in which species? Nanopublications from Wikidata
In December I reported about Groovy code to create nanopublications . This has been running for some time now, extracting nanopubs that assert that some metabolite is found in some species. I send the resulting nanopubs to Tobias Kuhn , to populate his Growing Resource of Provenance-Centric Scientific Linked Data (doi:10.1109/eScience.2018.00024, PDF).