Migrating pKa data from DrugMet to Wikidata
In 2010 Samuel Lampa and I started a pet project: collecting pKa data: he was working on RDF extension of MediaWiki and I like consuming RDF data. We started DrugMet. When you read this post, this MediaWiki installation may already be down, which is why I am migrating the data to Wikidata. Why? Because data curation takes effort, I like to play with Wikidata (see this H2020 proposal by Daniel Mietchen et al.), I like Open Data, and it still much needed.
We opted for a page with the minimal amount of information. To maximize the speed at which we could add information. However, when it came to semantics, we tried to be as explicit as possible, and, e.g. use the CHEMINF ontology. So, it collected:
- InChIKey (used to show images)
- the paper it was collected from (identified by a DOI)
- the value, and where possible, the experimental error
A page typically looks something like this:
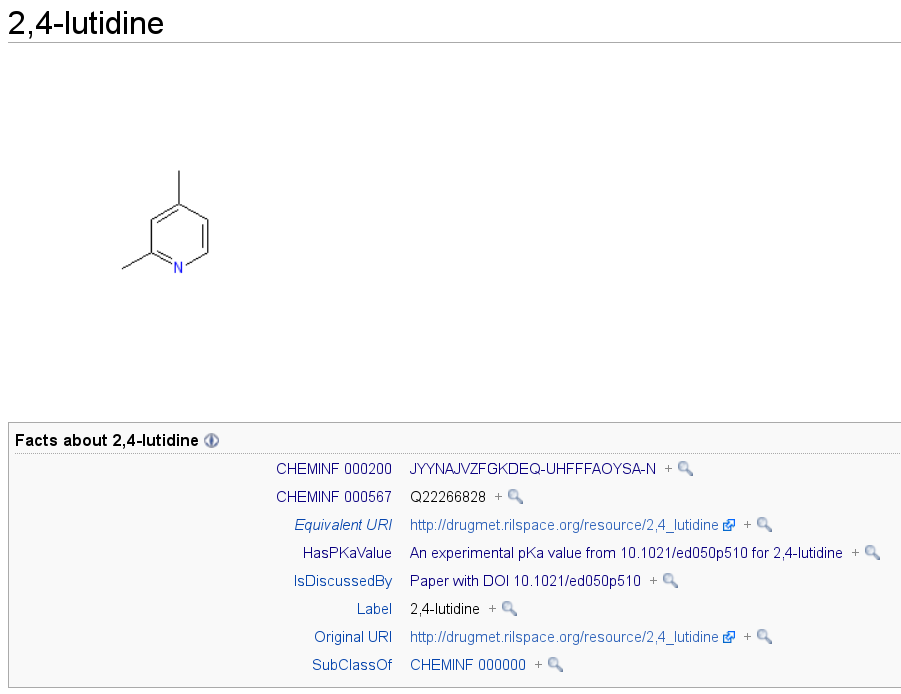
While not used on all pages, at some point I even started using templates, and I used these two, for molecules and papers:
{{Molecule
|Name=
|InChIKey=
|DOI=
|Wikidata=
}}
{{Paper
|DOI=
|Year=
|Wikidata=
}}
These templates, as well as the above screenshot, already contain a spoiler, but more about that later. Using MediaWiki functionality it was now easy to make lists, e.g. for all pKa data (more spoilers):
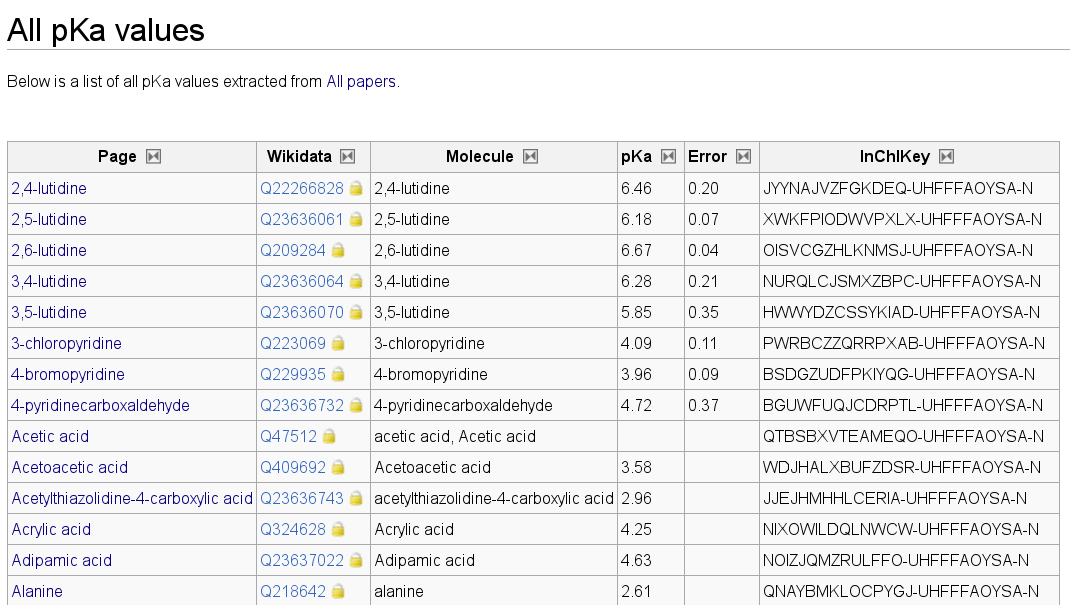
I find a database like this very important. It does not capture all the information it should be capturing, though, as is clear from the proposal some of use worked on a while back. However, this project got on hold; I don’t have time for it anymore, and it is not core to our department enough to spend time on write grant proposals for it.
But I still do not want to get this data get lost. Wikidata is something I have started using, as it is a machine readable CCZero database with an increasing amount of scientific knowledge. More and more people are working on it, and you must absolutely read this paper about this very topic (by a great team you should track, anyway). I am using it myself as source of identifier mappings and more. So, migrating the previously collected data to Wikidata makes perfect sense to me:
- if a compound is missing, I can easily create a new one using Bioclipse
- if a paper is missing, I can easily create a new one using Magnus Manske’s QuickStatements
- Wikidata has a pretty decent provenance model
I can annotate data with the data source (paper) it came from and also experimental conditions:
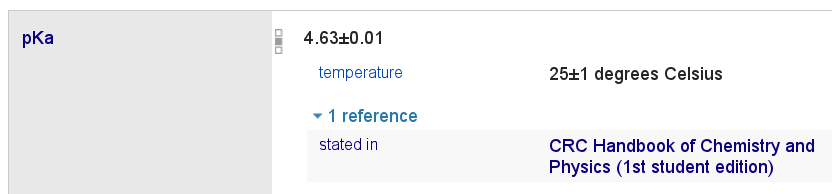
In fact, you’ll note that the the book is a separate Wikidata entry in itself. Better even, it’s an ‘edition’ of the book. This is the whole point we make in the above linked H2020 proposal: Wikidata is not a database specific for one domain, it works for any (scholarly) domain, and seamlessly links all those domains.
Now, to keep track of what data I have migrated, I am annotating DrugMet entries with links to Wikidata: everything with a Wikidata Q-code is already migrated. The above pKa table already shows Q-identifiers, but I also created them for all data sources I have used (three of them are two books and one old paper without a DOI):
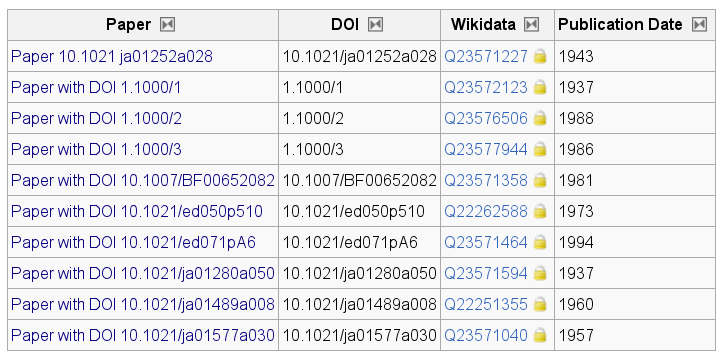
I have still quite a number of entries to do, but all the protocols are set up now.
On the downstream side, Wikidata is also great because of their SPARQL end point. Something that I did not get worked out some weeks ago, I did manage yesterday (after some encouragement from @arthursmith): list all pKa statements, including literature source if available:
If you run that query on the Wikidata endpoint, you get a table like this:
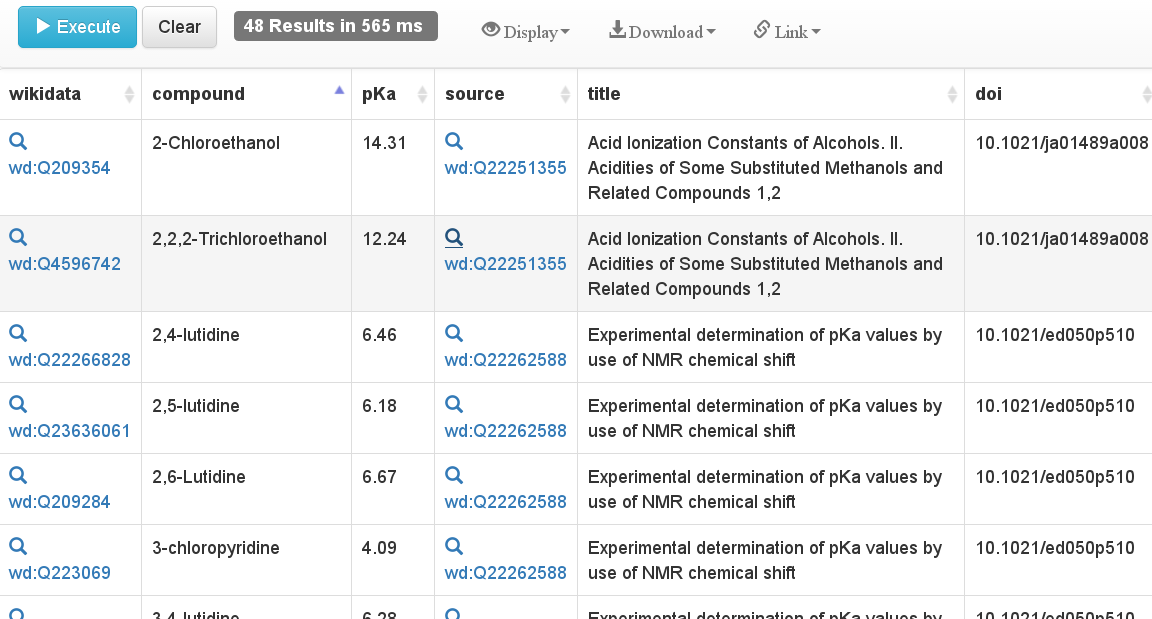
We here see experimental data from two papers: 10.1021/ja01489a008 and 10.1021/ed050p510. This can all be displayed a lot fancier, like make histograms, tables with 2D drawings of the chemical structures, etc, but I leave that to the reader.