-
CDK 2.12
Version 2.12 of the Chemistry Development Kit has been released. It is the last release with contributions by our NWO Open Science grant. This release adds some nice new APIs: -
Rescuing Scholia #3: We did it!
It was not a set up, when I openly wondered if we would be able to rescue Scholia in time. I honestly did not know. Three weeks and some serious hacking by an international team later I was more optimistic. Actually, just before christmas, we started writing a SWAT4HCLS 2026 demonstration abstract. This was accepted and you can read the Scholia 2026: Compliance with SPARQL 1.1 preprint here and here. This paper describes the work that had to be done, and I am deeply grateful to everyone who contributed with smaller or bigger contributions (Daniel, Peter, Konrad, Johannes, Lars, Wolfgang, Hannah). I am merely first author for the demo, and just another contributor to the long series of patches, in a branch started by Prof. Hannah Bast. -
Where do the WikiPathways come from?
WikiPathways was founded in 2008, in the year I left Wageningen (and we Nijmegen) and moved to Uppsala, Sweden. When we dediced to move back to The Netherlands in 2012, I got to opportunity to join the Department of Bioinformatics (BiGCaT) and work on Open PHACTS. I had visited the group in March 2011 because I had a COST action workshop near Maastricht (about nanoQSAR) and the bioinformatics group did WikiPathways. -
The TDCC NES Col-Lab Retreat
Last autumn two TDCC projects started, FAIR4ChemNL (with the PeerTube channel and doi:10.61686/XVYQV45374) and FAIRify for metabolomics data (doi:10.61686/CSGIP04334). But I haven’t written much on either yet and what the role is our research group in these projects. -
Open Infrastructures #2: the SURF Fediverse
When I first started writing this post, I started writing up why scientific communication is important, but because I started explaining what needs improving, and what are underlying causes why change is not happening, it got dark pretty quickly. So, I deleted that essay again. Instead, let’s just enjoy the awesome and long list of solutions we have for scientific discourse. Readers of my blog can find many posts in the past 20 years about the diversification. One thing I will say before I move one, is a reply to the argument that journal-based peer review is essential to the quality of research: if the quality of your research is dependent on your peers, then please rethink why you are doing research. -
Chemical blogs history
Like many awesome internet phenomena, blogging started in the late nineties. Nature authors and editors recognized the effort early. In 2006 there were already more than 45 million blogs, and at least 50 science blogs made it in the top 50,000 and 5 in the top 3,500. -

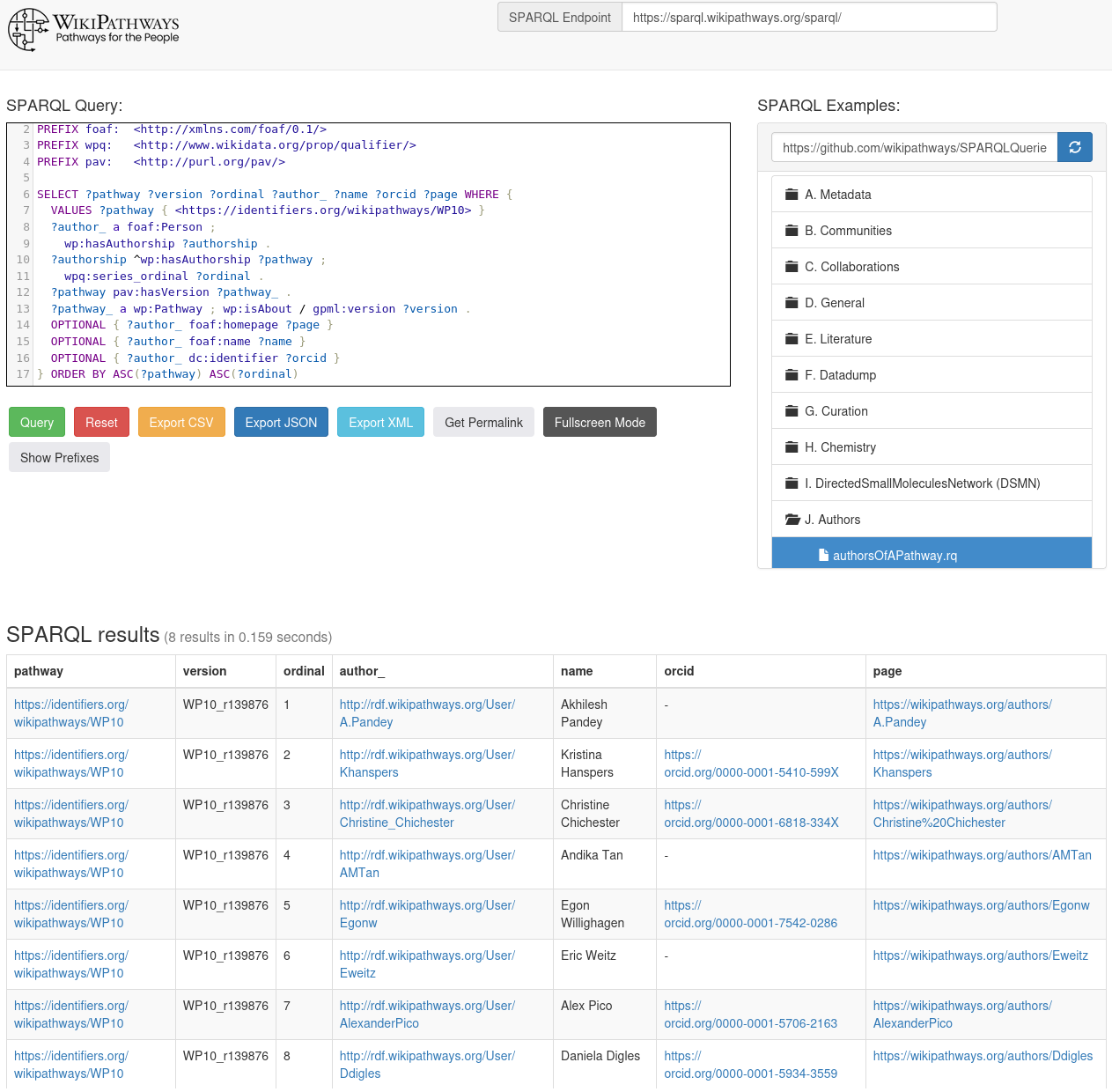
Where does the WikiPathways Cited In information come from?
I have been wanting to blog about this since this summer, but with everything going on, I never really got around to it. What is this Cited In feature of WikiPathways and where does that information come from? If you have not noticed this yet, this is what it looks like for WP4846:
- •
- 1
- 2