-
BridgeDb NWO grant update #1: first steps
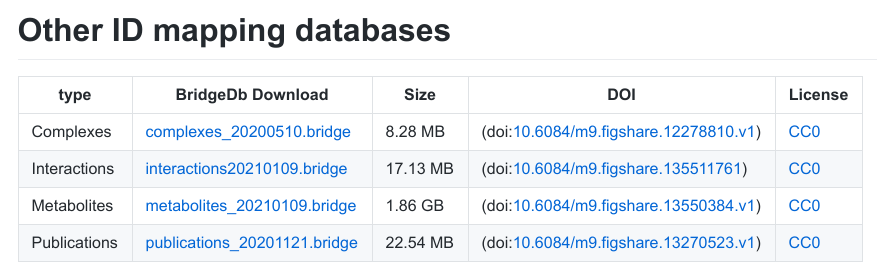
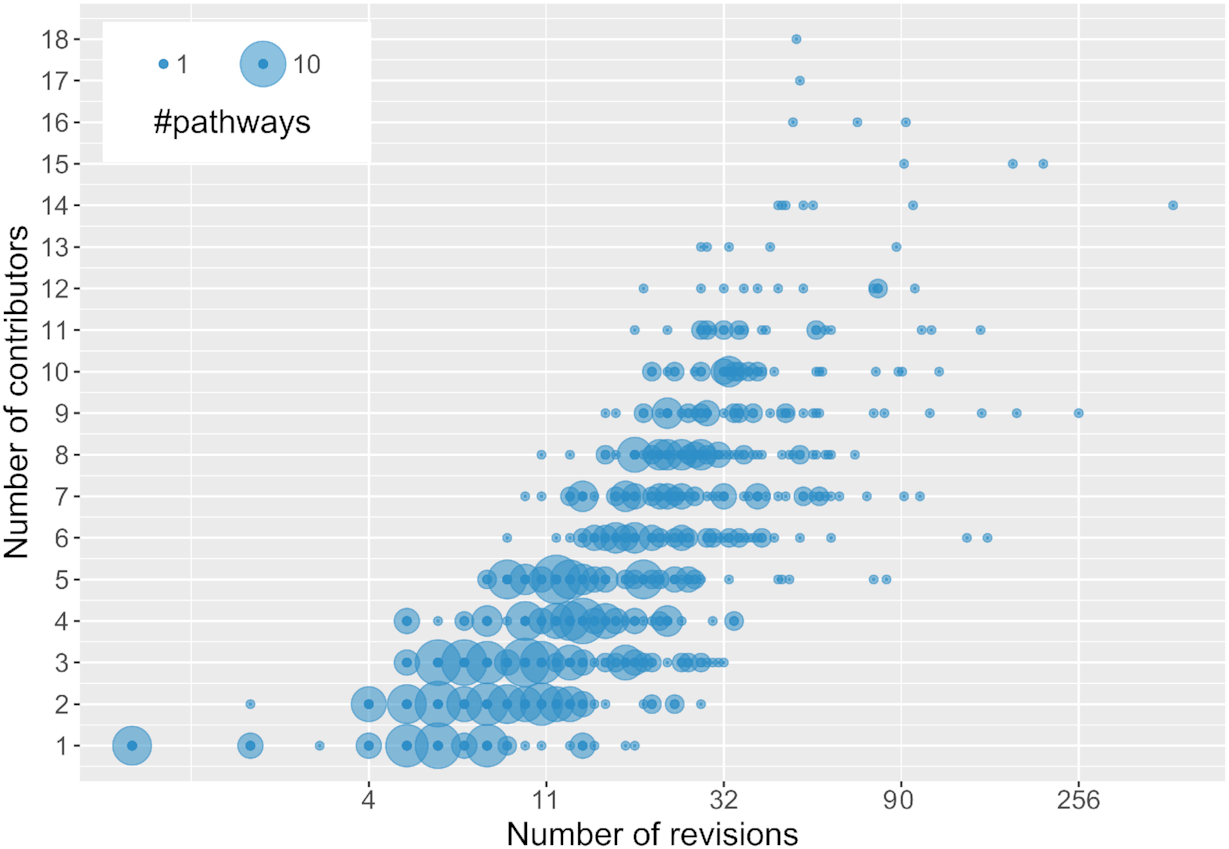
Last year, Denise, Tina, Marvin, and I received an NWO Open Science grant (203.001.121) to improve the long running BridgeDb project, originally developed by Martijn van Iersel (see doi:10.1186/1471-2105-11-5). Helena joined our group as research software engineer and will work part-time on this grant. We started two weeks ago, so time for an update of results: -

BioHackathon Europe 2021 #1: CiTO annotations in BioHackrXiv
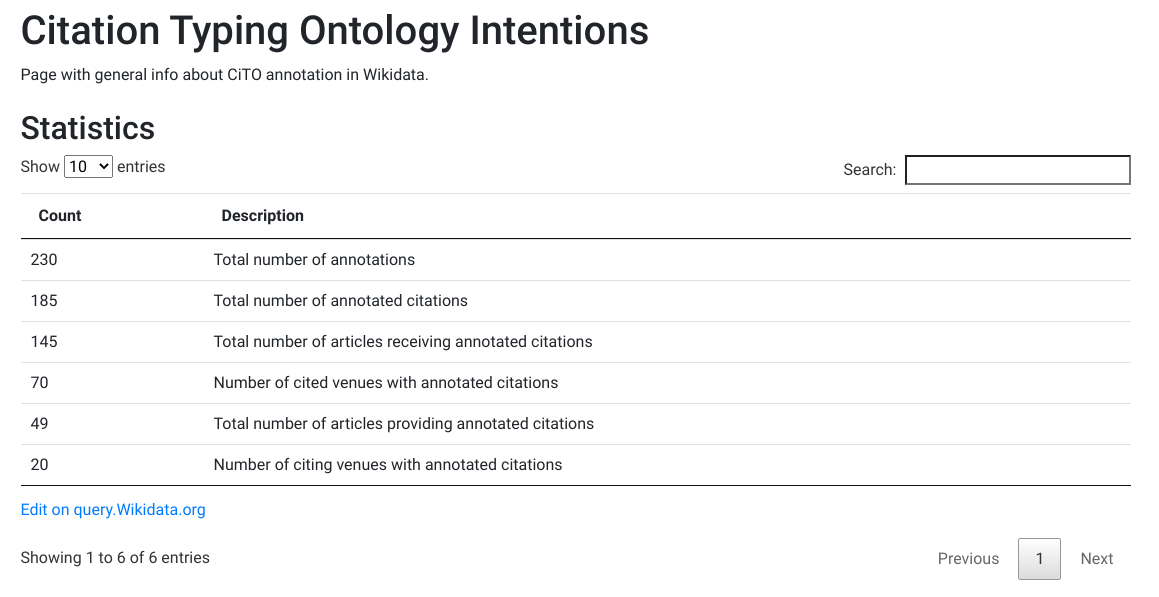
Serendipity. I did not plan this hack at the BioHackathon Europe 2021 but it happened anyway. Based on earlier work in the Journal of Cheminformatics, extending on the work by Krewinkel et al. I looked into the idea of using the Lua filter for BioHackrXiv, a preprint server for BioHackathons. Actually, I started by looking at the Citation Styling Language file used by the BioHackrXiv tools. But that was just wrong. -
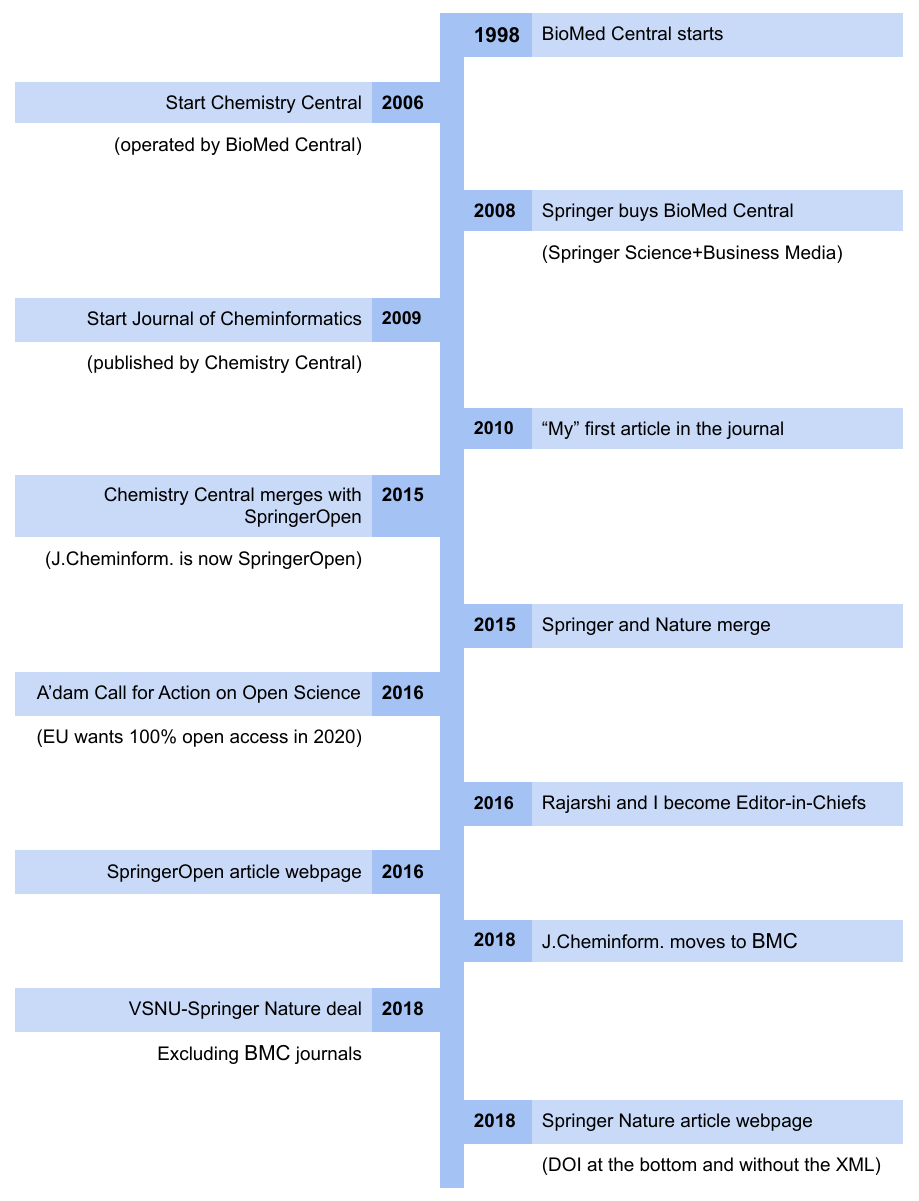
Conflict of Interest. Or why I am stepping down as Editor-in-Chief of the Journal of Cheminformatics.
Rough timeline of the Journal of Cheminformatics. The linked PDF has linked years with references. In this open letter, I will explain why I intend to step down as Editor-in-Chief of the Journal of Cheminformatics, which also happens to be a Springer Nature journal. It took me two years to come to this decision, and it cannot be claimed that I did not carefully evaluate the various aspects of it. However, I have now come to the conclusion that the opportunity it gives me to implement my ambition to shape open science chemistry now conflicts with the interests of Springer Nature. I will here outline some of the things I have taken into consideration.