-
Is your research cited by a Nobel prize winner?
Forget the journal impact factor and the H-index. You want your research being used. A first approximation of that is getting cited, sure. So, with the Nobel Prize week over (congrats to all winners! the Neanderthaler prize actually helped my work in Maastricht this week), let’s figure out of you are cited by a Nobel Prize winner. Wikidata allows us to figure this out with a SPARQL query (created together with Adriano): -
Wikidata now escapes SMILES and CXSMILES!
In the end it was a very simple change today (huge thanks to Nikki!), but Wikidata now escapes SMILES and CXSMILES (P10718) with the formatter URL (P1630)! -
Biology, ACPs, lipids, cheminformatics, and Dagstuhl
Already 3 months ago I visited Dagstuhl for the second time. The weather was much better than in the January right before the start of the pandemic. The first I attended the Computational Metabolomics meeting, with the focus From Cheminformatics to Machine Learning, one of the things we concerned ourselves with was how to do computation with compound classes (see Section 3.6 and this online book). We know how to handle SMILES and we know how to the substructure searching with SMARTS, but what if you have compound classes or lipid classes? Biology is a greasy business. -
new: "CAS Common Chemistry in 2021: Expanding Access to Trusted Chemical Information for the Scientific Community"
Open Science is happening. The merits are no longer theoretical or idealistic but tangible. Research is faster than ever, more vetted than ever (think PubPeer), more cited than ever. Fairly, not just because of Open Science, but open access causes readership causes impact causes citations. When new people and organizations start adopting Open Science this warms my hearth. -
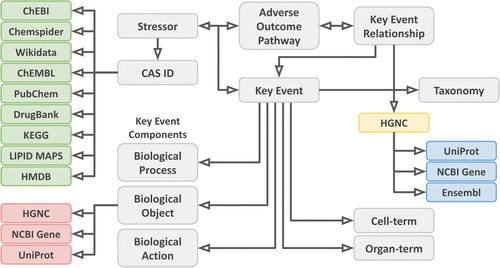
new: "Providing Adverse Outcome Pathways from the AOP-Wiki in a Semantic Web Format to Increase Usability and Accessibility of the Content"
I am a bit behind with tweeting about new published papers, but let that not reflect that these papers are not very exciting. The first paper is by Marvin an almost-finished PhD candidate in our group and now working as postdoc on the VHP4Safety project. He has been working on linking adverse outcome pathways (AOPs) with molecular pathways, such as in WikiPathways. This work was mostly done as part of the EU projects OpenRiskNet and EUToxRisk , during which he disseminated his research in many directions (e.g. the second paper in this post). Talking about impact. -

The International Conference on Chemical Structures scientific program is online!
Part of the scientific program of the ICCS 2022. Now that most speakers confirmed their talk by registering for the conference, it was time to upload the preliminary scientific program of the International Conference on Chemical Structures (#2022ICCS ).