ChemCuration: a small trick to fix the SMILES of glucuronides
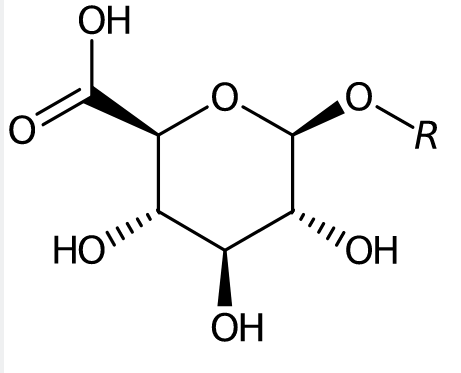
Glucuronide functional group.
Now that the ChemCuration 2019 online poster conference is nearing, and my upcoming talks about chemistry in Wikidata (also needing curation), and the much longer process of curation of metabolite (-like) structures in WikiPathways, I decided that something I tweeted earlier this week is actually quite useful, and therefore something I should really write up in my lab notebook.
Glucuronide is an example (biological) functional group. And there are several databases that represent the stereochemistry now always correct. That is an interoperability (and thus FAIR) problem. Correcting this is not trivial, particularly if you have to redraw the same glucuronide group again and again.
So, not looking forward to that, I invested a bit of time to find a SMILES trick. What if I had a SMILES snippet that I could easily copy/paste and attach to the SMILES of the chemical structure it is attached to? Here goes.
O1[C@H](C(O)=O)[C@H]([C@H](O)[C@@H](O)[C@@H]1O9)O.
I just realized that the original 3 I used can better be
a 9, which is less likely to occur in the SMILES of the rest of the molecule. The period at the end is also deliberate.
That way, I can just copy past the SMILES of the rest directly after that period. Then I get a disconnected structure, but
I only have to put a 9 next to the atom that is binding to the glucuronide. So, let’s see the R group is methane, I get:
O1[C@H](C(O)=O)[C@H]([C@H](O)[C@@H](O)[C@@H]1O9)O.C9
Now, next stop: CoA and other common biological tags.