Compound (class) identifiers in Wikidata
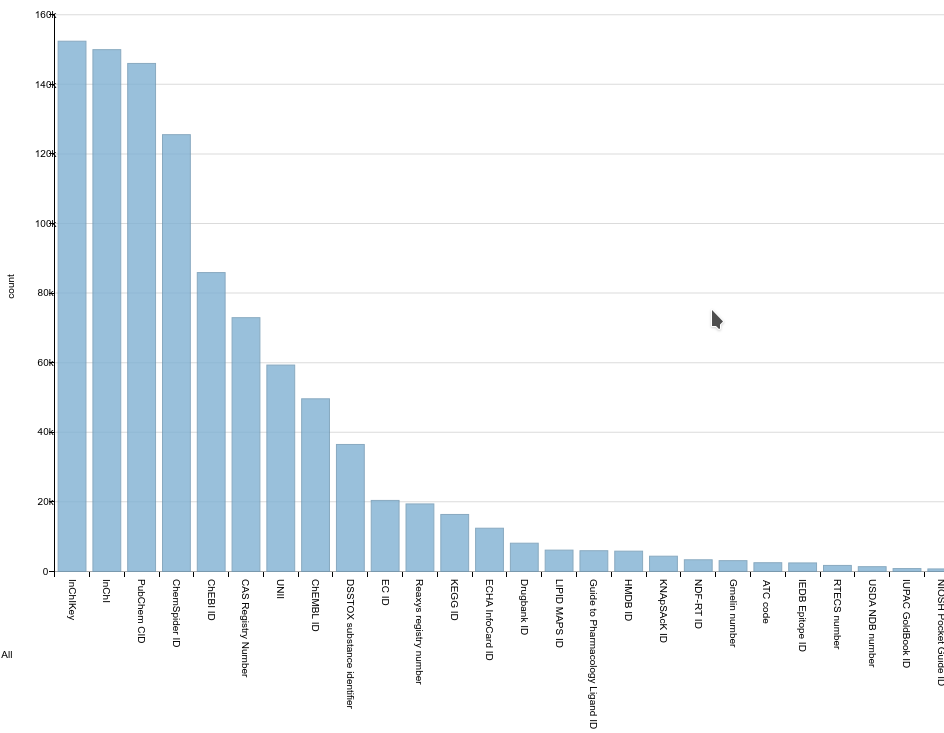
Bar chart showing the number of compounds with a particular chemical identifier.
I think Wikidata is a groundbreaking project, which will have a major impact on science. One of the
reasons is the open license (CCZero), the very basic approach (Wikibase), and the superb community around
it. For example, setting up your own Wikibase including a cool SPARQL endpoint, is
easily done with Docker.
Wikidata has many sub projects, such as WikiCite, which captures the collective of primary literature. Another one is the WikiProject Chemistry. The two nicely match up, I think, making a public database linking chemicals to literature (tho, very much needs to be done here), see my recent ICCS 2018 poster (doi:10.6084/m9.figshare.6356027.v1, paper pending).
But Wikidata is also a great resource for identifier mappings between chemical databases, something we need for our metabolism pathway research. The mapping, as you may know, are used in the latter via BridgeDb and we have been using Wikidata as one of three sources for some time now (the others being HMDB and ChEBI). WikiProject Chemistry has a related ChemID effort, and while the wiki page does not show much recent activity, there is actually a lot of ongoing effort (see plot). And I’ve been adding my bits.
Limitations of the links
But not each identifier in Wikidata has the same meaning. While they are all classified as ‘external-id’, the actual link may have different meaning. This, of course, is the essence of scientific lenses, see this post and the papers cited therein. One reason here is the difference in what entries in the various databases mean.
Wikidata has an extensive model, defined by the aforementioned WikiProject Chemistry. For example, it has different concepts for chemical compounds (in fact, the hierarchy is pretty rich) and compound classes. And these are differently modeled. Furthermore, it has a model that formalizes that things with a different InChI are different, but even allows things with the same InChI to be different, if need arises. It tries to accurately and precisely capture the certainty and uncertainty of the chemistry. As such, it is a powerful system to handle identifier mappings, because databases are not clear, and chemistry and biological in data is even less: we measure experimentally a characterization of chemicals, but what we put in databases and give names, are specific models (often chemical graphs).
That model differs from what other (chemical) databases use, or seem to use, because not always do databases indicate what they actually have in a record. But I think this is a fair guess.
ChEBI
ChEBI (and the matching ChEBI ID) has entries for chemical classes (e.g. fatty acid) and specific compounds (e.g. acetate).
PubChem, ChemSpider, UniChem
These three resources use the InChI as central asset. While they do not really have the concept of compound classes so much (though increasingly they have classifications), they do have entries where stereochemistry is undefined or unknown. Each one has their own way to link to other databases themselves, which normally includes tons of structure normalization (see e.g. doi:10.1186/s13321-018-0293-8 and doi:10.1186/s13321-015-0072-8).
HMDB
HMDB (and the matching P2057) has a biological perspective; the entries reflect the biology of a chemical. Therefore, for most compounds, they focus on the neutral forms of compounds. This makes linking to/from other databases where the compound is not neutral chemically less precise.
CAS registry numbers
CAS (and the matching P231) is pretty unique itself, and has identifiers for substances (see Q79529), much more than chemical compounds, and comes with a own set of unique features. For example, solutions of some compound, by design, have the same identifier. Previously, formaldehyde and formalin had different Wikipedia/Wikidata pages, both with the same CAS registry number.
Limitations of the links #2
Now, returning to our starting point: limitations in linking databases. If we want FAIR mappings, we need to be as precise as possible. Of course, that may mean we need more steps, but we can always simplify at will, but we never can have a computer make the links more complex (well, not without making assumptions, etc).
And that is why Wikidata is so suitable to link all these chemical databases: it can distinguish differences when needed, and make that explicit. It make mappings between the databases more FAIR.