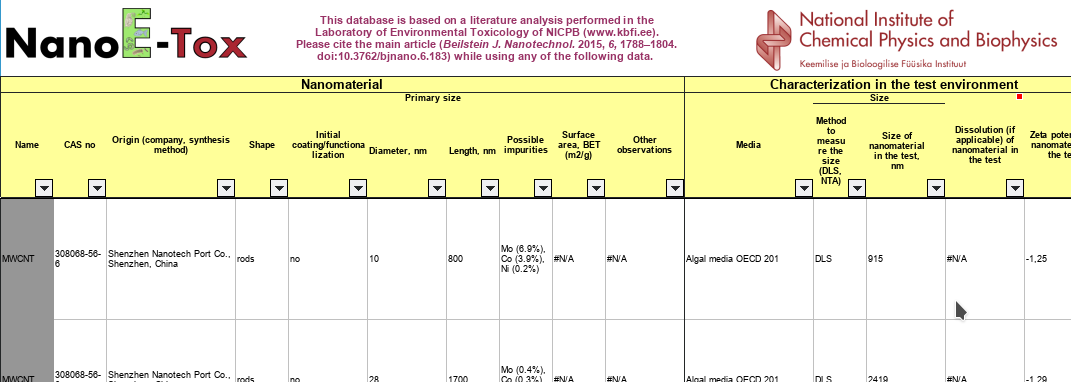
-

What metabolites are found in which species? Nanopublications from Wikidata
In December I reported about Groovy code to create nanopublications . This has been running for some time now, extracting nanopubs that assert that some metabolite is found in some species. I send the resulting nanopubs to Tobias Kuhn , to populate his Growing Resource of Provenance-Centric Scientific Linked Data (doi:10.1109/eScience.2018.00024, PDF). -
Join me in encouraging the ACS to join the Initiative for Open Citations
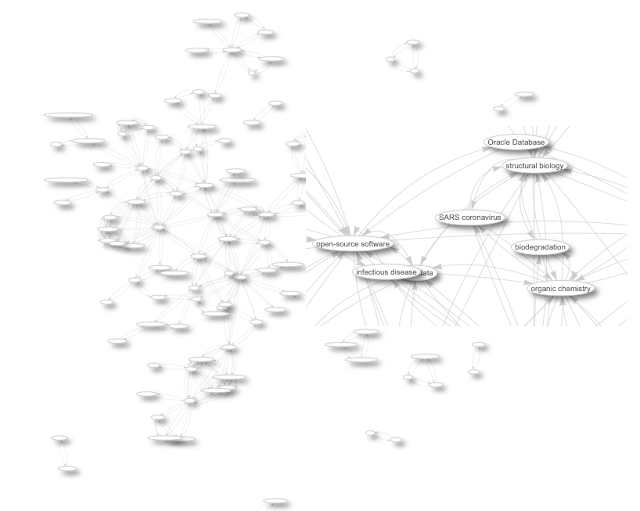
My research is into abstract representation of chemical information, important for other research to be performed. Indeed, my work is generally reused, but knowing which research fields my work is used in, or which societal problems it is helping solve, is not easily retrieved or determined. Efforts like WikiCite and Scholia do allow me to navigate the citation network, so that I can determine which research fields my output influences and which diseases are studied with methods I proposed. Here’s a network of topics of articles citing my work: -
Programming in the Life Sciences #23: research output for the future
A random public domain picture with 10 in it. Ensuring that you and others can understand you research output five years from now requires effort. This is why scholars tend to keep lab notebooks. The computational age has perhaps made us a bit lazy here, but we still make an effort. A series of Ten Simple Rules articles outline some of the things to think about: