-
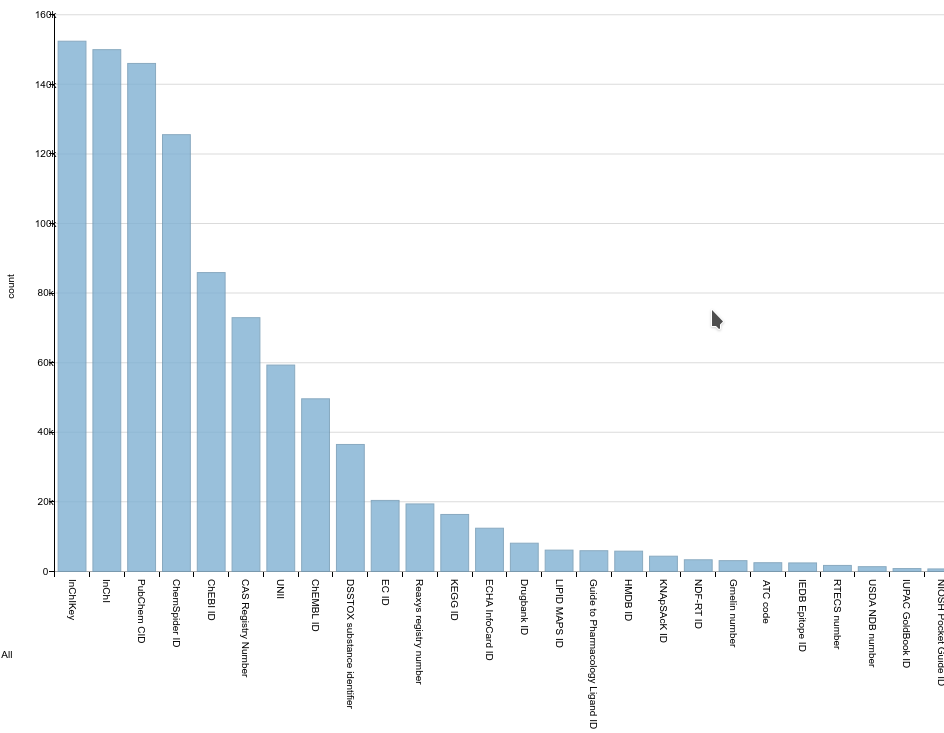
Compound (class) identifiers in Wikidata
Bar chart showing the number of compounds with a particular chemical identifier. I think Wikidata is a groundbreaking project, which will have a major impact on science. One of the reasons is the open license (CCZero), the very basic approach (Wikibase), and the superb community around it. For example, setting up your own Wikibase including a cool SPARQL endpoint, is easily done with Docker. -
New paper: "Integration among databases and data sets to support productive nanotechnology: Challenges and recommendations"
The U.S.A and European nanosafety communities have a longstanding history of collaboration. On both sides there are working groups, NanoWG and WG-F (previously called WG4) of the NanoSafety Cluster. I have been chair of WG4 for about three years and still active in the group, though in the past half year, without dedicated funding, less active. That is already changing again with the imminent start of the NanoCommons project. -
Two conference proceedings: nanopublications and Scholia
It takes effort to move scholarly publishing forward. And the traditional publishers have not all shown to be good at that: we’re still basically stuck with machine-broken channels like PDFs and ReadCubes. They seem to all love text mining, but only if they can do it themselves.
.jpg)