-
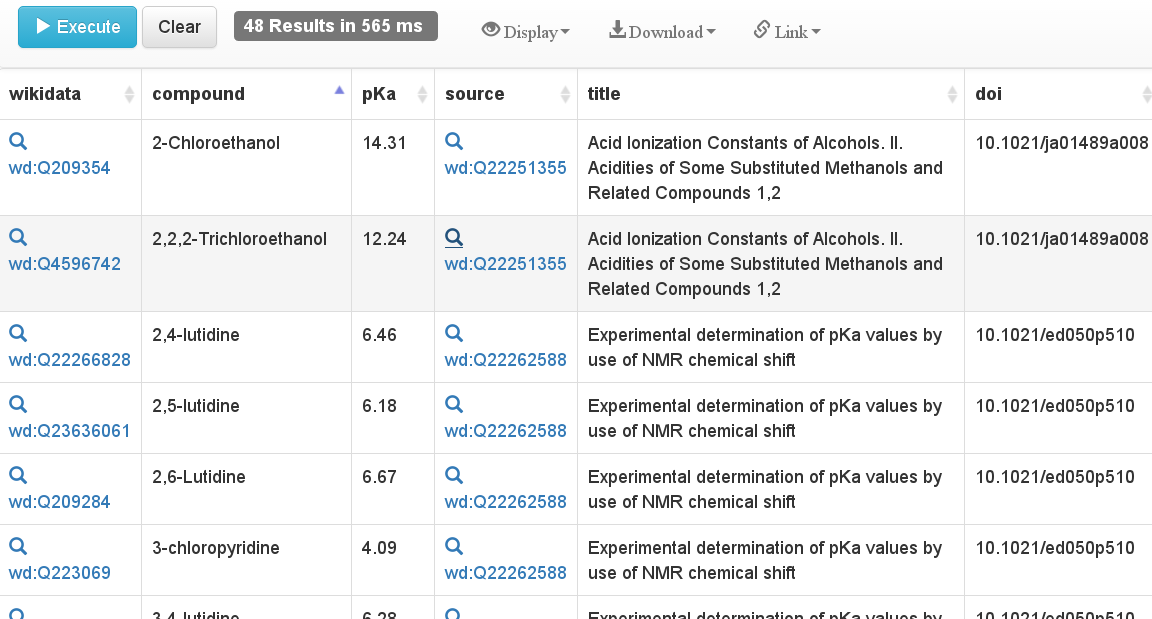
Migrating pKa data from DrugMet to Wikidata
In 2010 Samuel Lampa and I started a pet project: collecting pKa data: he was working on RDF extension of MediaWiki and I like consuming RDF data. We started DrugMet. When you read this post, this MediaWiki installation may already be down, which is why I am migrating the data to Wikidata. Why? Because data curation takes effort, I like to play with Wikidata (see this H2020 proposal by Daniel Mietchen et al.), I like Open Data, and it still much needed. -
Adding disclosures to Wikidata with Bioclipse
Last week the huge, bi-annual ACS meeting took place (#ACSSanDiego), during which commonly new drug (leads) are disclosed. This time too, like this one tweeted by Bethany Halford: -
Adding chemical compounds to Wikidata
Adding chemical compounds to Wikidata is not difficult. You can store the chemical formula (P274), (canonical) SMILES (P233), InChIKey (P235) (and InChI (P234), of course), as well various database identifiers (see what I wrote about that here ]). It also allows storing of the provenance, and has predicates for that too. -

Coding an OWL ontology in HTML5 and RDFa
There are many fancy tools to edit ontologies. I like simple editors, like nano. And like any hacker, I can hack OWL ontologies in nano. The hacking implies OWL was never meant to be hacked on a simple text editor; I am not sure that is really true. Anyways, HTML5 and RDFa will do fine, and here is a brief write up. This post will not cover the basics of RDFa and does assume you already know how triples work. If not, read this RDFa primer first. -
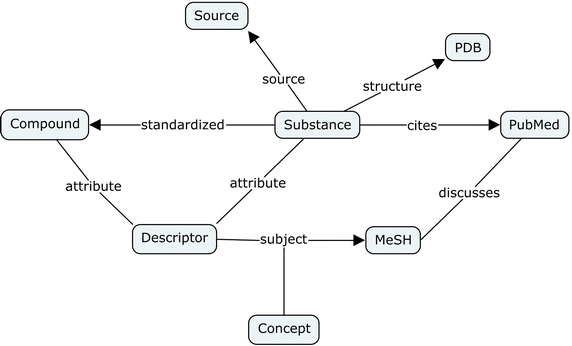
PubChemRDF: semantic web access to PubChem data
Gang Fu and Evan Bolton have blogged about it previously, but their PubChemRDF paper is out now (doi:10.1186/s13321-015-0084-4). It very likely defines the largest collection of RDF triples using the CHEMINF ontology and I congratulate the authors with a increasingly powerful PubChem database. -

Chemistry Central and the ORCID identifier
If you are a scientist you have heard about the ORCID identifier by now. If not, you have been focusing on groundbreaking research and isolated yourself from the rest of the world, just to make it perfect and get that Nobel prize next year. If you have been working on impactful research, Nobel prize-worthy, and have been blogging and tweeting about your progress, as a good Open Scholar, you know ORCID is the DOI for “research contributors” and you already have one yourself, and probably also that T-shirt with your own identifier. Mine is 0000-0001-7542-0286, and almost 1.3M other authors got one too. The list of ORCIDs on Wikipedia is growing (and Wikidata), thanks to Andy Mabbett, whom also made it possible to add your ORCID on WikiPathways.