Programming in the Life Sciences #17: The Open PHACTS scientific questions
I think the authors of the Open PHACTS proposal made a right choice in defining a small set of questions that the solution to be developed could be tested against. The questions being specific, it is much easier to understand the needs. In fact, I suspect it may even be a very useful form of requirement analysis, and makes it hard to keep using vague terms.
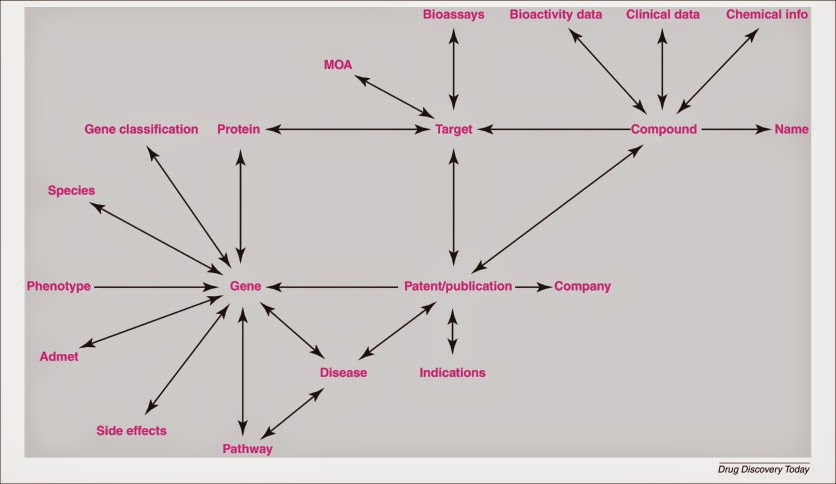
Open PHACTS has come up with 20 questions (doi:10.1016/j.drudis.2013.05.008; Open Access):
- Give me all oxidoreductase inhibitors active <100 nM in human and mouse
- Given compound X, what is its predicted secondary pharmacology? What are the on- and off-target safety concerns for a compound? What is the evidence and how reliable is that evidence (journal impact factor, KOL) for findings associated with a compound?
- Given a target, find me all actives against that target. Find/predict polypharmacology of actives. Determine ADMET profile of actives
- For a given interaction profile – give me similar compounds
- The current Factor Xa lead series is characterized by substructure X. Retrieve all bioactivity data in serine protease assays for molecules that contain substructure X
- A project is considering protein kinase C alpha (PRKCA) as a target. What are all the compounds known to modulate the target directly? What are the compounds that could modulate the target directly? I.e. return all compounds active in assays where the resolution is at least at the level of the target family (i.e. PKC) from structured assay databases and the literature
- Give me all active compounds on a given target with the relevant assay data
- Identify all known protein–protein interaction inhibitors
- For a given compound, give me the interaction profile with targets
- For a given compound, summarize all ‘similar compounds’ and their activities
- Retrieve all experimental and clinical data for a given list of compounds defined by their chemical structure (with options to match stereochemistry or not)
- For my given compound, which targets have been patented in the context of Alzheimer’s disease?
- Which ligands have been described for a particular target associated with transthyretin-related amyloidosis, what is their affinity for that target and how far are they advanced into preclinical/clinical phases, with links to publications/patents describing these interactions?
- Target druggability: compounds directed against target X have been tested in which indications? Which new targets have appeared recently in the patent literature for a disease? Has the target been screened against in AZ before? What information on in vitro or in vivo screens has already been performed on a compound?
- Which chemical series have been shown to be active against target X? Which new targets have been associated with disease Y? Which companies are working on target X or disease Y?
- Which compounds are known to be activators of targets that relate to Parkinson’s disease or Alzheimer’s disease
- For my specific target, which active compounds have been reported in the literature? What is also known about upstream and downstream targets?
- Compounds that agonize targets in pathway X assayed in only functional assays with a potency <1 μM
- Give me the compound(s) that hit most specifically the multiple targets in a given pathway (disease)
- For a given disease/indication, give me all targets in the pathway and all active compounds hitting them
Students in the Programming in the Life Sciences course will this year pick one of these questions as a starting point in the project. The goal is to develop a HTML+JavaScript solution that will answer the question the selected. There is freedom to tweak the question to personal interests, of course. By selecting a simpler pharmacological question that last year, more time and effort can be put into visualization and interpretation of the found data.