Two Apache Jena SPARQL query performance observations
Doing searches in RDF stores is commonly done with SPARQL queries. I have been using this with the semantic web translation of WikiPathways by Andra to find common content issues, though sometimes combined with some additional Java code. For example, find PubMed identifiers that are not numbers.
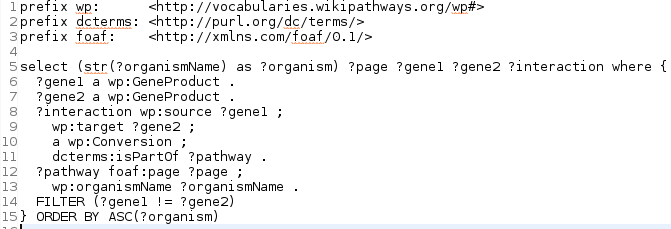
Based on Ryan’s work on interactions, a more complex curation query I recently wrote in reply to issues that Alex ran into with converting pathways to BioPax, is to find interactions that convert a gene to another gene. Such occurred in WikiPathways because graphically you do not see the difference. I originally had this query:
SELECT (str(?organismName) as ?organism) ?page
?gene1 ?gene2 ?interaction
WHERE {
?gene1 a wp:GeneProduct .
?gene2 a wp:GeneProduct .
?interaction wp:source ?gene1 ;
wp:target ?gene2 ;
a wp:Conversion ;
dcterms:isPartOf ?pathway .
?pathway foaf:page ?page ;
wp:organismName ?organismName .
} ORDER BY ASC(?organism)
This query properly found all gene-gene conversions to be fixed. However, it was also horribly slow with my JUnit/Apache Jena set up. The queries runs very efficiently on the Virtuoso-based SPARQL end point. I had been trying to speed it up in the past, but without much success. Instead, I ended up batching the testing on our Jenkins instance. But this got a bit silly, with at some point subsets of less than 100 pathways.
Observation #1
So, I turned to twitter, and quite soon got
three useful
leads. The first two suggestions did not help, but helped me rule out the problem.
Of course, there is literature about optimizing, like this recent paper by Antonis (doi:10.1016/j.websem.2014.11.003),
but I haven’t been able to convert this knowledge into practical steps either. After ruling out these options (though I kept the
sameTerm() suggestion), and realized it had to be the first two triples with the variables ?gene1 and ?gene2. So,
I tried using FILTER there too,
resulting with this query:
WHERE {
?interaction wp:source ?gene1 ;
wp:target ?gene2 ;
a wp:Conversion ;
dcterms:isPartOf ?pathway .
?pathway foaf:page ?page ;
wp:organismName ?organismName .
FILTER (!sameTerm(?gene1, ?gene2))
FILTER (?gene1 a wp:GeneProduct)
FILTER (?gene2 a wp:GeneProduct)
} ORDER BY ASC(?organism)
That did it! The time to run a query halved. Not so surprising, in retrospect, but it all depends on the SPARQL engine: which parts does it run first. Apparently, Jena’s SPARQL engine starts at the top. This seems to be confirmed by the third comment I got. However, I always understood engine can also start at the bottom.
Observation #2
But that’s not all. This speed up made me wonder something else. The problem clearly seems to engine approach to run parts of the query. So, what if I remove further choices in what to run first? That leads me to a second observation. It helps significantly if you reduce the number of subgraphs it should later “merge”. Instead, if possible, use property paths. That again, about halved the runtime of the query. I ended up with the below query, which, obviously, no longer give me access to the pathway resources, but I can live with that:
WHERE {
?interaction wp:source ?gene1 ;
wp:target ?gene2 ;
a wp:Conversion ;
dcterms:isPartOf/foaf:page ?pathway ;
dcterms:isPartOf/wp:organismName ?organismName .
FILTER (!sameTerm(?gene1, ?gene2))
FILTER EXISTS {?gene1 a wp:GeneProduct}
FILTER EXISTS {?gene2 a wp:GeneProduct}
} ORDER BY ASC(?organism)
I’m hoping these two observations may help other with using Apache Jena with unit and integrated testing of RDF generation too.