-
Two conference proceedings: nanopublications and Scholia
It takes effort to move scholarly publishing forward. And the traditional publishers have not all shown to be good at that: we’re still basically stuck with machine-broken channels like PDFs and ReadCubes. They seem to all love text mining, but only if they can do it themselves. -
New Paper: "Using the Semantic Web for Rapid Integration of WikiPathways with Other Biological Online Data Resources"
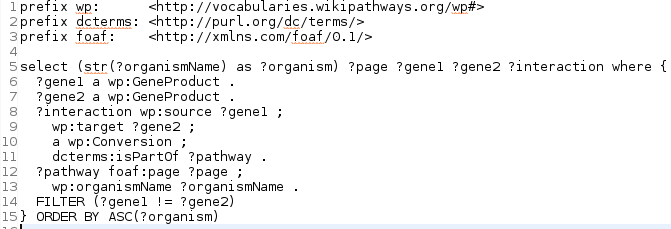
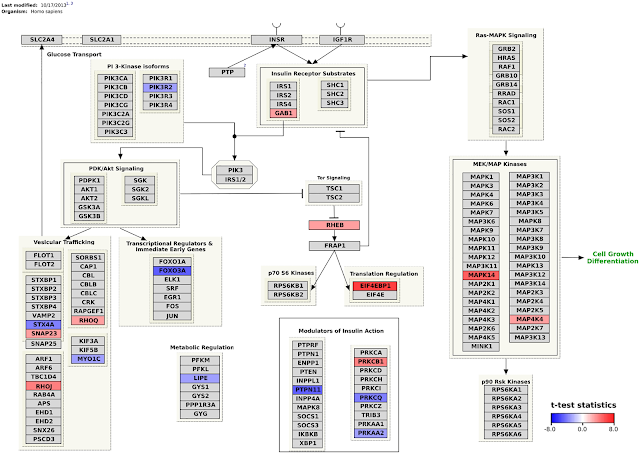
Andra Waagmeester published a paper on his work on a semantic web version of the WikiPathways (doi:10.1371/journal.pcbi.1004989). The paper outlines the design decisions, shows the SPARQL endpoint, and several examples SPARQL queries. These include federates queries, like a mashup with DisGeNET (doi:10.1093/database/bav028) and EMBL-EBI’s Expression Atlas. That results in nice visualisations like this: -
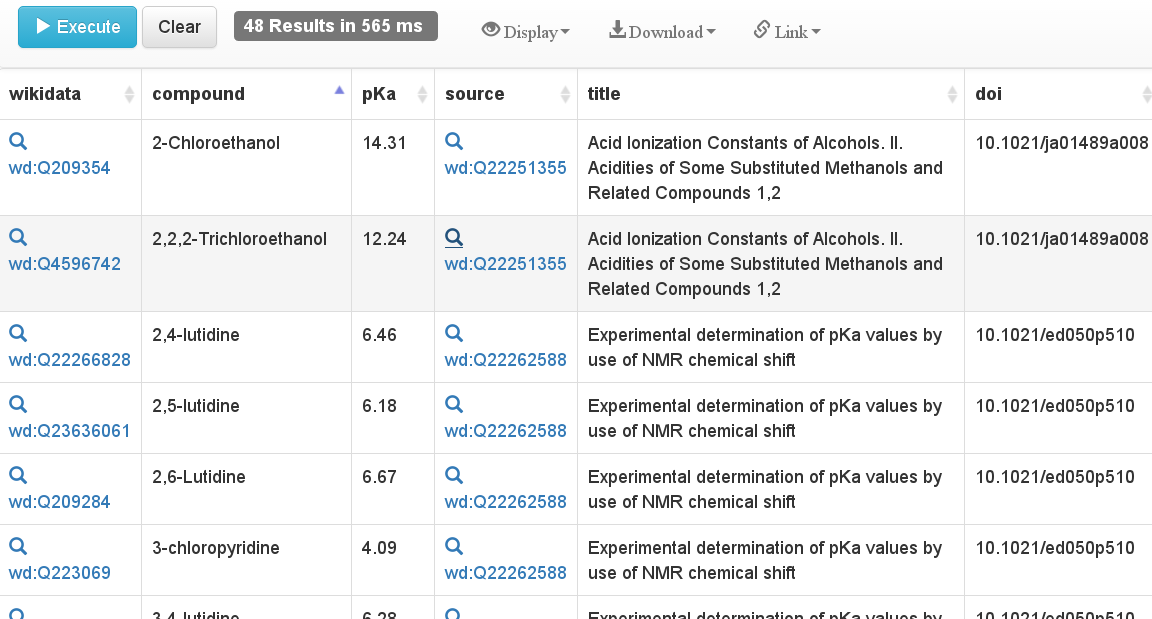
Migrating pKa data from DrugMet to Wikidata
In 2010 Samuel Lampa and I started a pet project: collecting pKa data: he was working on RDF extension of MediaWiki and I like consuming RDF data. We started DrugMet. When you read this post, this MediaWiki installation may already be down, which is why I am migrating the data to Wikidata. Why? Because data curation takes effort, I like to play with Wikidata (see this H2020 proposal by Daniel Mietchen et al.), I like Open Data, and it still much needed. -
Adding disclosures to Wikidata with Bioclipse
Last week the huge, bi-annual ACS meeting took place (#ACSSanDiego), during which commonly new drug (leads) are disclosed. This time too, like this one tweeted by Bethany Halford:
.jpg)