-
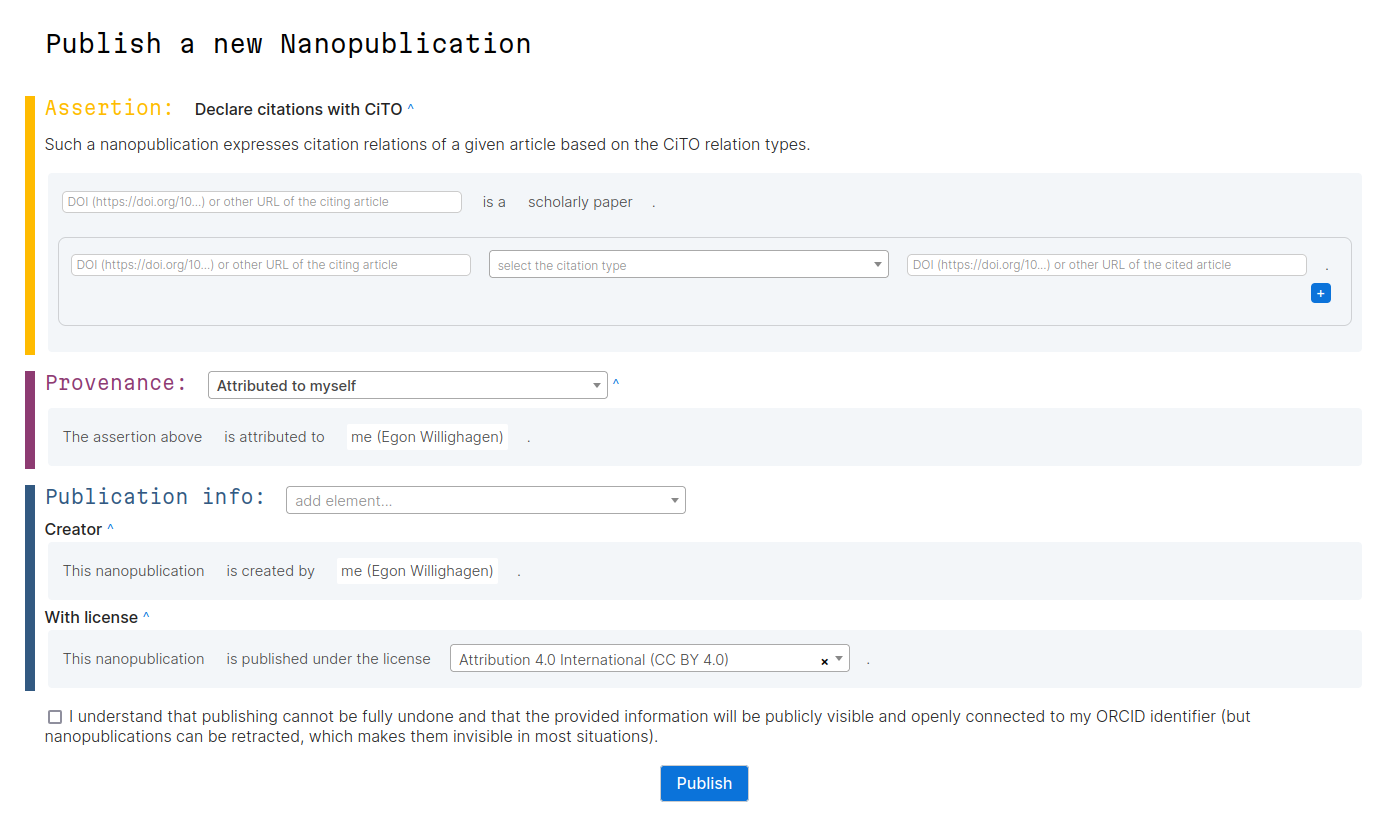
Open Science Retreat #2: CiTO Nanopublications
During the Open Science Retreat I organized a short session where we looking into typing citation intentions using a new nanopublication template. First, let’s describe nanopublications (originally used in doi:10.3233/ISU-2010-0613) a bit. Scholia gives a nice overview of (macro?)publications on the topic. The nanopub.net website describes that [a nanopublication is a small knowledge graph snippet with metadata that is treated as an independent (scientific) publication.]. The knowledge graph, it continues, can be anything from an opinion to the link between a disease and a gene (doi:10.1109/ESCIENCE.2018.00024). -
Open Science Retreat #1: impressions
Last week I attended the Open Science Retreat (#osr24nl) in a quite and relaxing region in North-Holland. The meeting was how I like all meetings to be (and I count myself lucky many of my meetings are like this): open, welcoming, constructive, diverse, and intellectually challenging. Not all scientific meetings are like this and it is easy to end up going to obligatory meetings where the discussions are of a different level. Therefore, great thanks to the organizers, but also to all participants, that showed not just to have a hearth for open science (getting pretty common), but also a drive to advocate for open science. Finally, I like to thank the people that joined me in creating nanopublications for CiTO annotations (will blog about that later), and to Sadik and Marija with whom we worked on exploring using Wikibase for capturing knowledge about research waste in ecology (more about that later too). -
Reusing data: two new papers
My research is about the interaction of (machine) representation and the impact on the success of data analysis (matchine learning, chemometrics, AI, etc). See the posts about molecular chemometrics. This got me into FAIR: making data interoperable and being able to (really) reuse data is the starting point of doing research. -
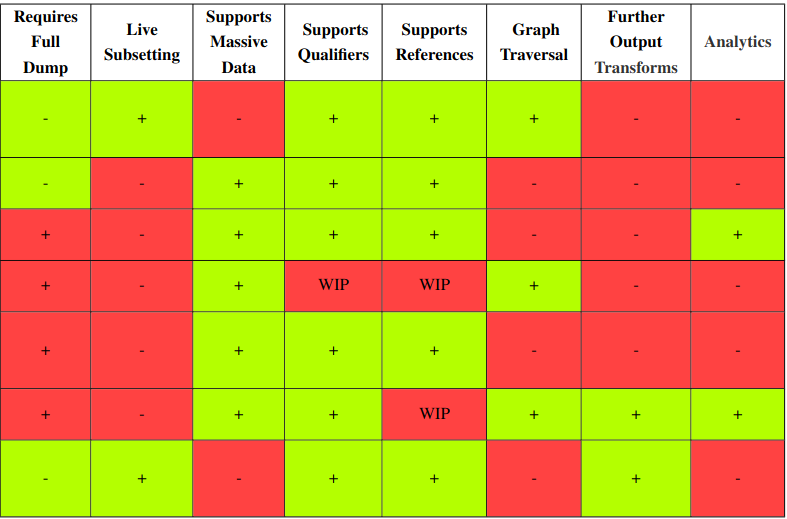
New paper: "Wikidata subsetting: approaches, tools, and evaluation"
Just before the end of the year, the Wikidata subsetting: approaches, tools, and evaluation paper by Seyed Amir Hosseini Beghaeiraveri et al. got published (doi:10.3233/SW-233491). I am really excited our group (i.e. Ammar and Denise) has been able to contribute to this. I think it also is a great example of the power of hackathons to bring together people. -
PhD Defences: Andra Waagmeester and Marvin Martens
2023 has been a long year in which a lot happens. Two EU projects ended (RiskGONE and NanoSolveIT; more about that in a later post), our group leader Chris Evelo will retire this year, the ELIXIR Toxicology Community started (see this post), the new WikiPathways website launched (see this post), and a lot, lot more. -
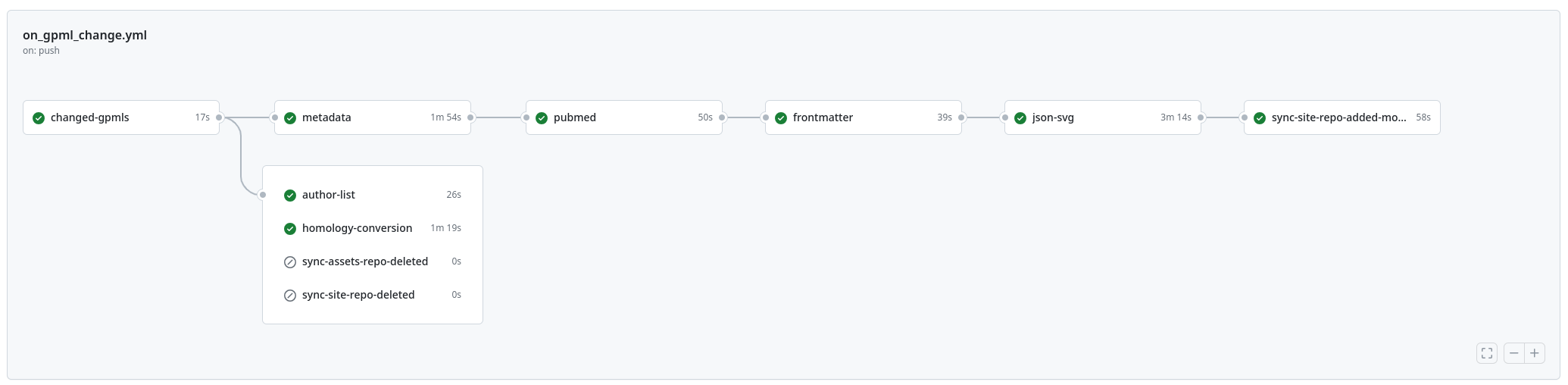
New paper: "WikiPathways 2024: next generation pathway database"
This week the next WikiPathways NAR Database issue paper was published (doi:10.1093/nar/gkad960). It is the next paper in a series of papers about the evolution of the Open Science project for making biological pathways available in a Open and FAIR way. This year, it described that significant move away from MediaWiki. It simply was too costly to keep up with the upstream code base (think: more than 200 thousand euro costly). This paper describes a transition to a modular system with Jekyll and Markdown as new platform technologies. The full details are available as open notebook science: everything is basically a git repository.