New paper: "WikiPathways 2024: next generation pathway database"
This week the next WikiPathways NAR Database issue paper was published (doi:10.1093/nar/gkad960). It is the next paper in a series of papers about the evolution of the Open Science project for making biological pathways available in a Open and FAIR way. This year, it described that significant move away from MediaWiki. It simply was too costly to keep up with the upstream code base (think: more than 200 thousand euro costly). This paper describes a transition to a modular system with Jekyll and Markdown as new platform technologies. The full details are available as open notebook science: everything is basically a git repository.
The is the workflow of what the new platform does when a new pathway (version) gets added to WikiPathways:
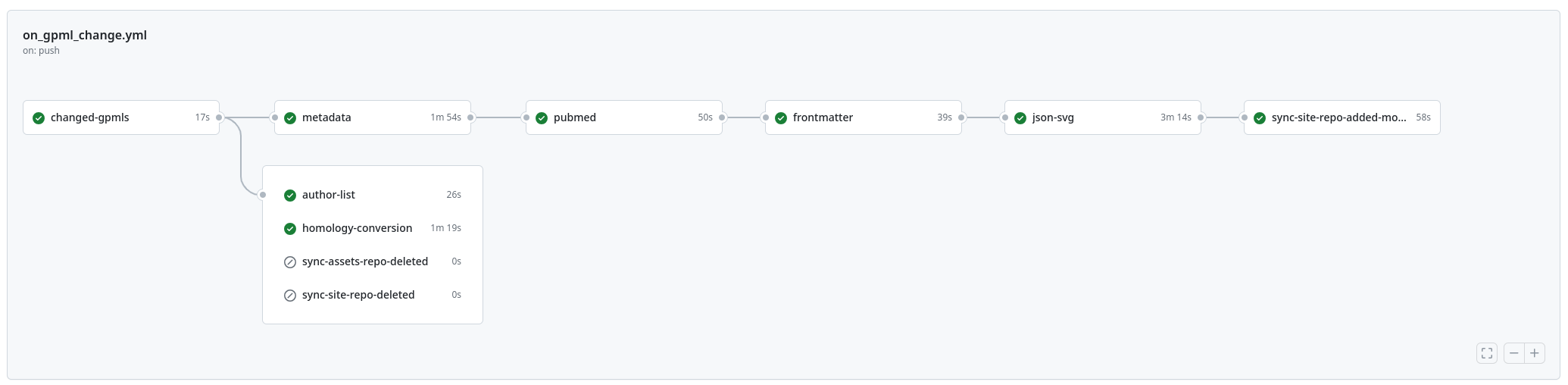
The upgrade of the whole stack is, however, in full swing. Not everything has migrated yet and the RDF generation is not for example.