-
The Molecular Chemometrics Principles #1: access to data
The meetings in and around Oxford were great! I already wrote that the Predictive Toxicology workshop was brilliant (see Oxford… #1 ) and Oxford… #2 ), but I also very, very much enjoyed meeting up with Dan and Nico! During the week, someone (name and address is know at the editorial office) commented on the fact that my blog posts are somewhat difficult to follow; that is, it’s often not clear why I am posting what I am posting. -
Oxford... #2
The Predictive Toxicology meeting is over. It was a great meeting, by any standard. Very much recommended, and many thanx to Barry for the organization! The meeting was a true workshop, with a mix of presentations and getting work done. I participated in a group that looked at mutagenicity of potential anti-malaria drugs from the datasets of GSK and Novartis recently release as Open Data. We used various tools to predict properties, and plan to make all our results freely available soon. Otherwise, it was also great to meet Nina again (with whom I talked about OpenTox), and to meet other CDK users, including Patrik (SMARTCyp , doi:10.1021/ml100016x) and David (Inkspot). -
Oxford...
Yesterday I arrived in Oxford, after a 3.5 hour bus transfer from London Stansted. Long, boring ride (though I might have seen a few red kites , but seeing that they were near extinct, I am wondering what other large bird of prey has strong split tail like a swallow). Showed once more that the UK infrastructure has hardly changed since the 19th century. Enjoying an undergraduate room at one of the colleges. Pretty basic, but makes me feel more like a human than a tourist. Yes!, undergraduate students are human too! One of the advantages is you get an excellent internet connection :) -
ChemPedia RDF #1: the SPARQL end point
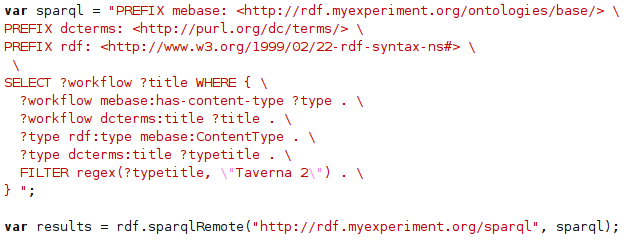
Well, you might spot a pattern here; yes, another chemical SPARQL end point (actually, it shares the end point with the Solubility data). This time around Rich’s ChemPedia. Taking advantage of the CC0-licensed downloads , I have created a small Groovy script (using this JSON library) to convert the ChemPedia JSON into Notation3: -
Really free chemistry books
With pleasure I read Analogue or Digital? - Both, Please. Funnily, I just created MP3 (or, preferably Ogg Vorbis, superior but hardly any support by commercial companies, who rather seem to pay license fees) directly from the CD.