-
Programming in the Life Sciences #3: the assessment
Now that I have wrote out the goals, what they students will practically do, and how to get started with the Open PHACTS platform, I will list how we will assess the students: -
Programming in the Life Sciences #2: accounts and API keys
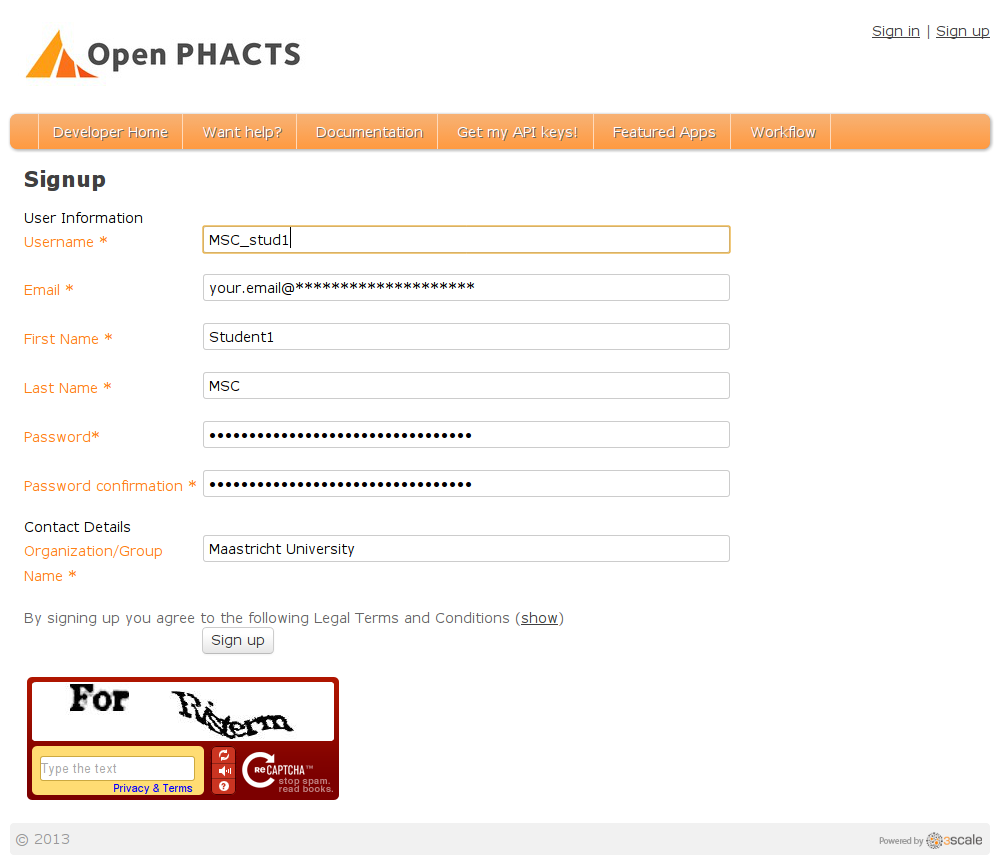
I have outlined the scope of the six-day course: the students will learn to program while hacking on the Open PHACTS’ Linked Data API (LDA). The first step is to get an account for the LDA. I have already done that to save time. But these are the steps to take. You go to https://dev.openphacts.org/signup: -
Programming in the Life Sciences #1: a six day course
Our department will soon start the course Programming in the Life Sciences for a group of some 10 students from the Maastricht Science Programme. This is the first time we give this course, and over the next weeks I will be blogging about this course. First, some information. These are the goals, to use programming to: -
CiteULike adds a HTML widget to embed citations
Spjuth, O.; Carlsson, L.; Alvarsson, J.; Georgiev, V.; Willighagen, E.; Eklund, M. Current Topics in Medicinal Chemistry 2012, 12, 1980-1986. -
"Emerging practices for mapping and linking life sciences data using RDF"
The “Emerging practices for mapping and linking life sciences data using RDF” (doi:10.1016/j.websem.2012.02.003) is now available online, where I contributed a section on the original workflow for creating ChEMBL triples, and contributed to the section about open licensing, referring to CCZero and the Panton Principles. Happy reading! -
Visualizing metabolite fluxes on WikiPathways pathways using a PathVisio plugin
Visualizing metabolite fluxes on WikiPathways pathways using a PathVisio plugin