-
Visualizing metabolite fluxes on WikiPathways pathways using a PathVisio plugin
Visualizing metabolite fluxes on WikiPathways pathways using a PathVisio plugin -
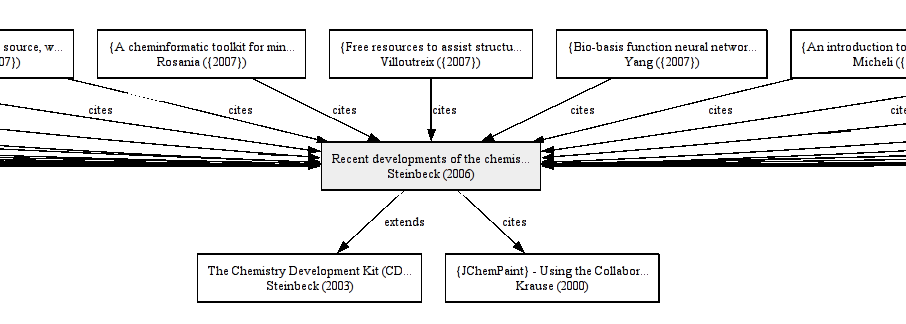
CiTO / CiteULike: publishing innovation
Readers of my blog know I have been using the Citation Typing Ontology, CiTO (doi:10.1186/2041-1480-1-S1-S6). I allows me to see how the CDK is cited and used . CiteULike is currently adding more CiTO more functionality, which they started doing almost one and a half years ago. -
Groovy Cheminformatics 4th edition
Six month was not quite the amount of time I anticipated between the third and fourth edition, but I finally managed to upload edition 1.4.7-0 of my Groovy Cheminformatics book. The first three editions sold 37 copies, including two for myself. Enough to feel supported and to continue working on it. -
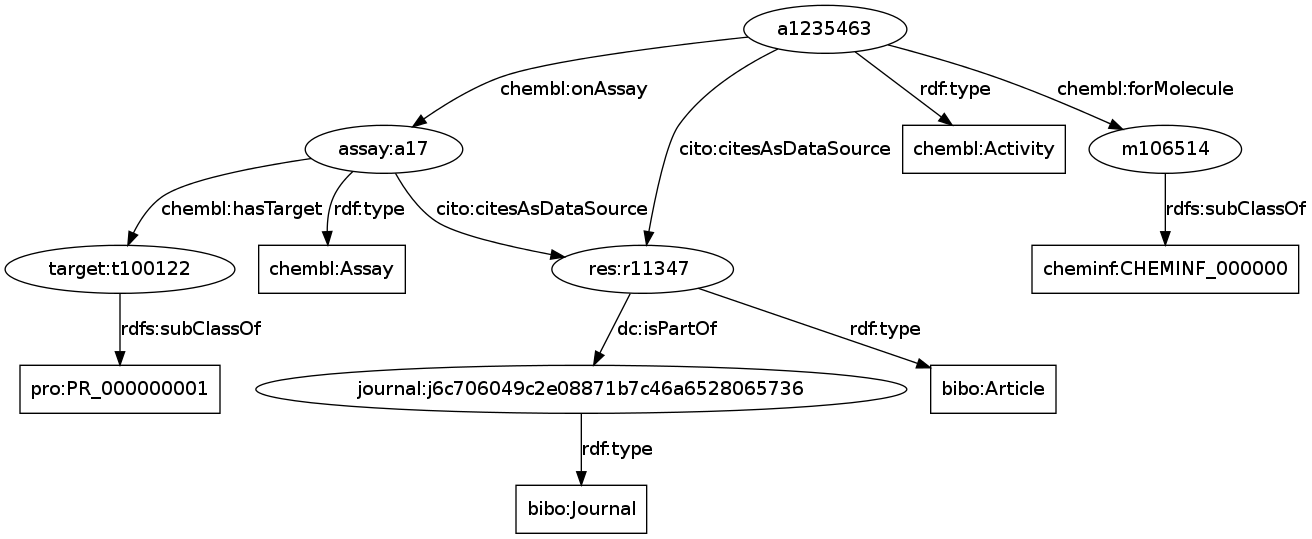
ChEMBL-RDF: Uploading data to Kasabi with pytassium
I reported earlier how to I uploaded the ChemPedia (RIP) data onto Kasabi. But for ChEMBL-RDF I have used the pytassium tool, not just because it has a cool name :) I discovered yesterday, however, that I did not write down in this lab notebook, what steps I needed to take to reproduce it. And I just wanted to uploaded new triples to the ChEMBL-RDF data set on Kasabi. -
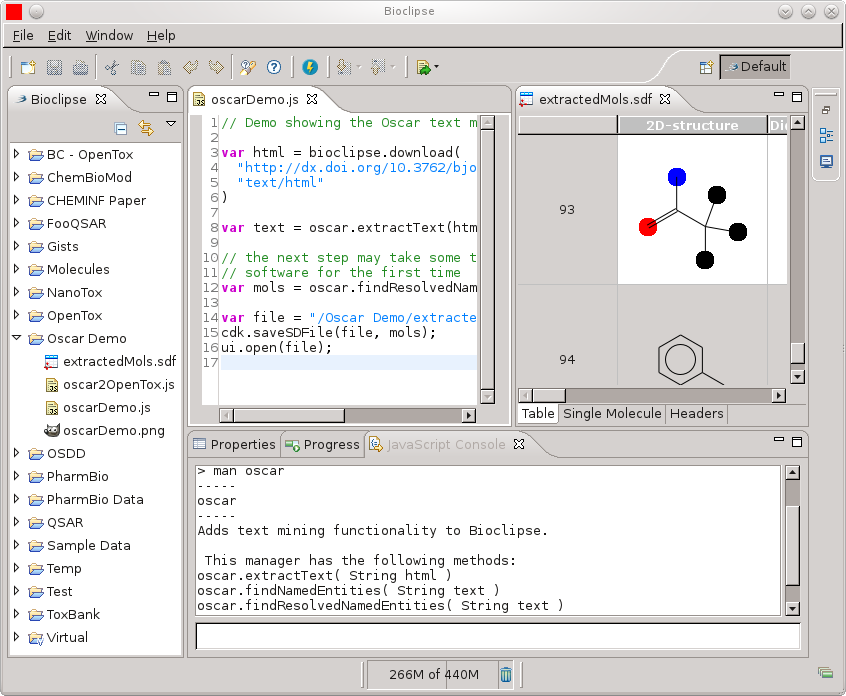
Bioclipse-Oscar4 - Text mining in Bioclipse
Almost a year ago I started a position with Peter Murray-Rust to work on Oscar for three months (see this overview of results; a paper by the full Oscar team (Sam, David, Dan, Lezan) is pending, and I’m really happy to have been able to contribute bits to the project). Since then, I have had little time :( That’s how it goes, with post-hopping, unfortunately. One thing I did do after that, was write a Bioclipse plugin.