Bioclipse and SPARQL end points #2: MyExperiment
RDF and SPARQL are two really useful Open Standards. Bioclipse-RDF is a plugin for Bioclipse that provide RDF functionality, among which using remote SPARQL end points.
The MyExperiment team has set up an excellent RDF front end. For example, this is my MyExperiment account in RDF. The storage gets updated once every day (at this moment), but I’m sure that will become more often in the future. The SPARQL end point allows us to make any query against the database that their ontologies support. The above query showed up 132 workflows when I ran it today.
Gists
Now, so far I have been using Gist to share Bioclipse scripts and I wrote some Bioclipse GUI elements for downloading such gists . To annotate these gists, Delicious has been used, and a listing of Bioclipse scripts can be found under the tags bioclipse and gist.
MyExperiment also allows to share workflows, but originally only for Taverna. A recent change, however, made it possible to share other types of workflows too. And, MyExperiment itself also allows all the annotation which we may want to do.
Now, using the Bioclipse-RDF functionality, I can query the MyExperiment database and use that information do to stuff. If this stuff is a Bioclipse script, then I can just download it, as the download link of a workflow is part of the RDF too, as we will see.
Querying a SPARQL end point
As we have seen in the first article of this series , the RDF manager his a method to query a remote SPARQL end point. The complexity is mostly in formulating the SPARQL (and this one happens to be available as workflow on MyExperiment too:
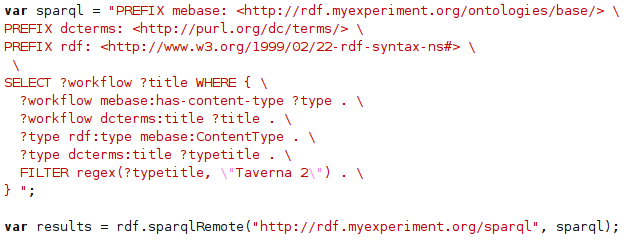
This is worsened by the fact that JavaScript does not have a type of multiline Strings, so the backslashes at the end of the lines are JavaScript syntax and not part of the SPARQL. To simplify the SPARQL, I will show below the SPARQL only, and not the Bioclipse script wrapping as is done in the above code snippet.
List all Taverna 2 workflows
Listing all Taverna 2 workflows, as shown in that earlier snippet, is done with the SPARQL:
This query asks for a ?workflow and its ?title, and the workflow ?type must be of Class ContentType as defined in the
mebase namespace, and we want to know the ?typetitle of that content type, because we are filtering that using a
regular expression to contain “Taverna 2”. Well, if you cannot
follow this, just google for SPARQL,
and run one of those tutorials which are abundantly present on the web.
Finding tags used to annotate workflows
To list all tags which have likely to do with metabolomics, I can do:
And I can also list all workflows that are tagged like this. Because I could not get string matching to work, I used the tag’s URI instead:
All MyExperiments Users in Sweden
I was also interested in all MyExperiment Users in Sweden, and again, a simple SPARQL tells me where they live:
Finding Duncan and Pierre
Very easy to find users, such as Duncan:
Or Pierre, who has not listed where he lives:
My workflows
Given a user, it is also easy to get the workflows he owns. Again, I am using my URI instead of combining with a search for my account, because the MyExperiment SPARQL end point is not particularly fast:
Earlier in this series: