-
Join me in encouraging the ACS to join the Initiative for Open Citations
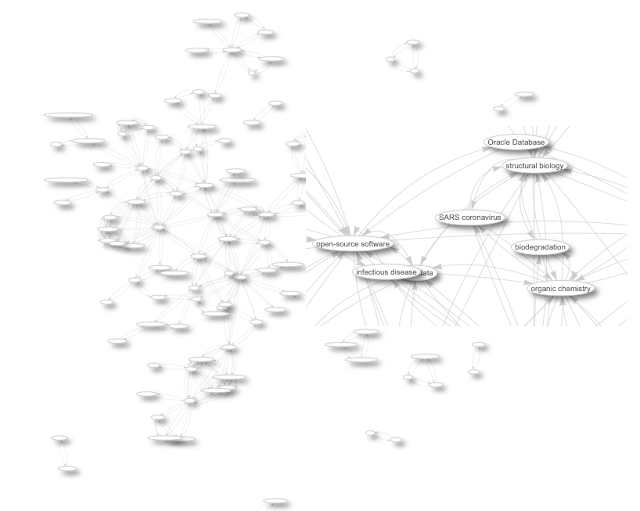
My research is into abstract representation of chemical information, important for other research to be performed. Indeed, my work is generally reused, but knowing which research fields my work is used in, or which societal problems it is helping solve, is not easily retrieved or determined. Efforts like WikiCite and Scholia do allow me to navigate the citation network, so that I can determine which research fields my output influences and which diseases are studied with methods I proposed. Here’s a network of topics of articles citing my work: -
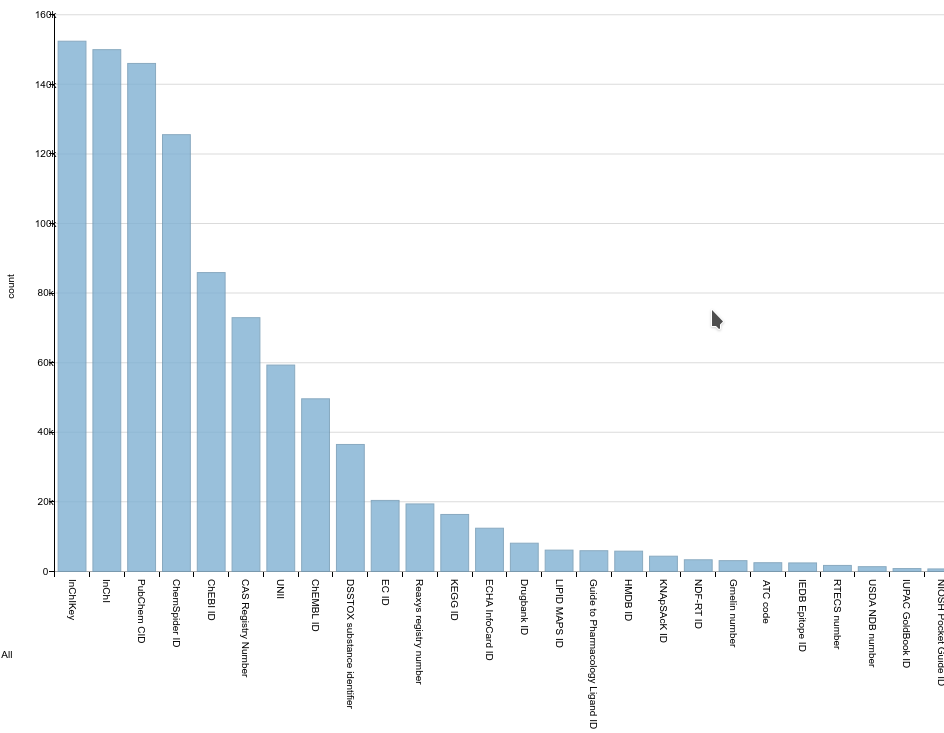
Compound (class) identifiers in Wikidata
Bar chart showing the number of compounds with a particular chemical identifier. I think Wikidata is a groundbreaking project, which will have a major impact on science. One of the reasons is the open license (CCZero), the very basic approach (Wikibase), and the superb community around it. For example, setting up your own Wikibase including a cool SPARQL endpoint, is easily done with Docker. -
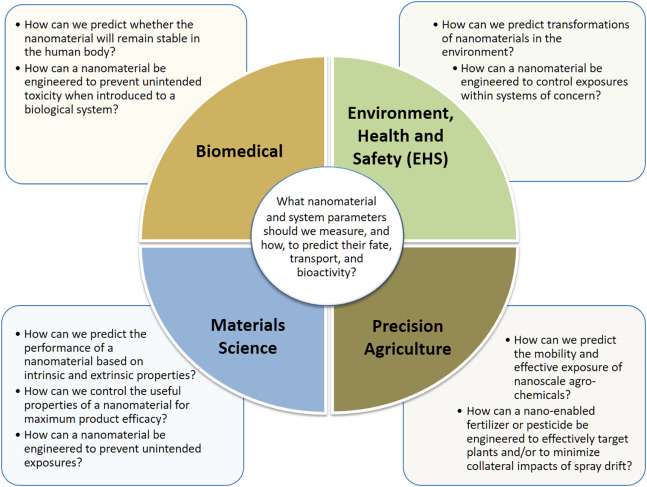
New paper: "Integration among databases and data sets to support productive nanotechnology: Challenges and recommendations"
The U.S.A and European nanosafety communities have a longstanding history of collaboration. On both sides there are working groups, NanoWG and WG-F (previously called WG4) of the NanoSafety Cluster. I have been chair of WG4 for about three years and still active in the group, though in the past half year, without dedicated funding, less active. That is already changing again with the imminent start of the NanoCommons project.

.jpg)