-

Archiving and updating my blog
This blog is almost 18 years old now. I have long wanted to migrate it to a version control system and at the same time have more control over things. Markdown would be awesome. In the past year, I learned a lot about the power of Jekyll and needed to get more experienced with it to use it for more databases, like we now do for WikiPathways. -
Universities and open infrastructures
The role of a university is manifold. Being a place where people can find knowledge and the track record how that knowledge was reached is often seen as part of that. Over the past decades universities outsources this role, for example to publishers. This is seeing a lot of discussion and I am happy to see that the Dutch Universities are taking back control fast now. For example, Radboud University (>1k followers) already joined the Fediverse (Mastodon etc), making them independent from non-EU law and commercial interests. Scientific journals, Nobel Prize winners, etc already joined too , btw. -
Journal Rankings
I am pleased to learn that the Dutch Universities start looking at rankings of a more scientific way. It is long overdue that we take scientific peer review of the indicators used in those rankings seriously, instead of hiding beyond fud around the decline of quality of research. -

Qeios, an open dissemination platform for research output
A bit over a year ago I got introduced to Qeios when I was asked to review an article by Michie, West, and Hasting: “Creating ontological definitions for use in science” (doi:10.32388/YGIF9B.2). I wrote up my thoughts after reading the paper, and the review was posted openly online and got a DOI. Not the first platform to do this (think F1000), but it is always nice to see some publishers taking publishing seriously. Since then, I reviewed two more papers. -
Twitter exits FAIR and is no longer a dissemination solution
And just like that, without a warning, Twitter changed policies again, and you now need a Twitter account and be logged in to see public tweets: Twitter has started blocking unregistered users (The Verge). Though I learned it first via Mastodon, of course. -
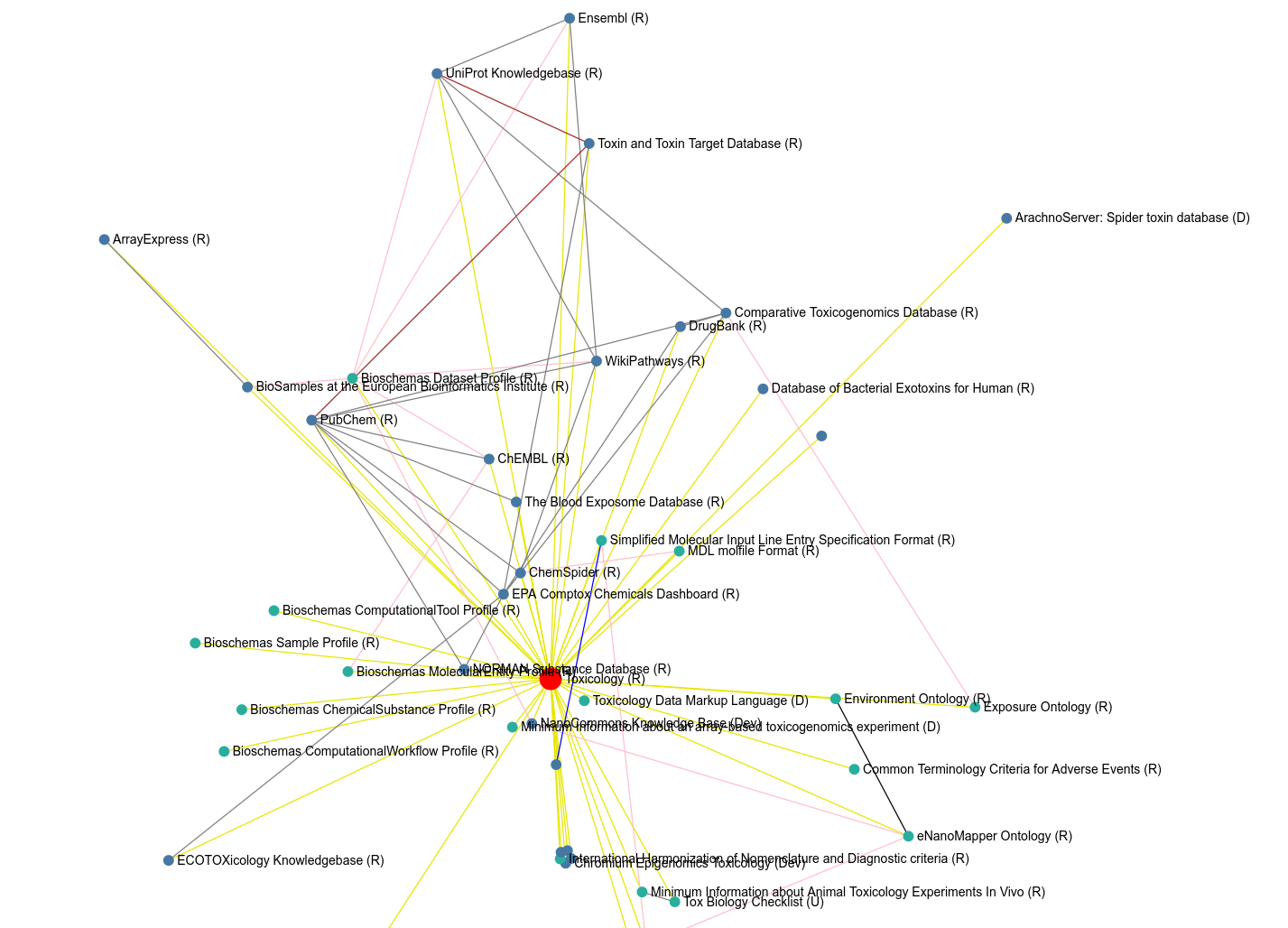
Community activity #2: FAIRsharing
Some years ago we started the ELIXIR Toxicology Community. It has been an interesting journey, partly covered in this whitepaper). We started with interaction we had in several projects already, but particularly the potential. I see this. This series of posts is a number of things toxicology projects can do to benefit from ELIXIR solutions (“services”). The posts have been sent first to the ELIXIR Toxicology Community mailing list (please join!). -
Information Retrieval versus ChatGPT
When last week in a large (and relevant) Dutch research event ChatGPT came up, and that this was going to change the world. Even the critiques came up, but were effectively disregarded with “these methods get better very quickly”. This is not untrue, but not really true either. I murmur “not even wrong”. I know how hard it is to get computers to find meaningful patters; I did a PhD in this in the early 21st century.