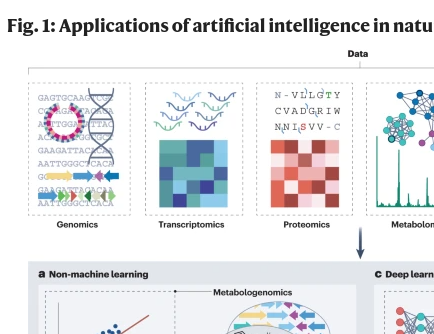
-
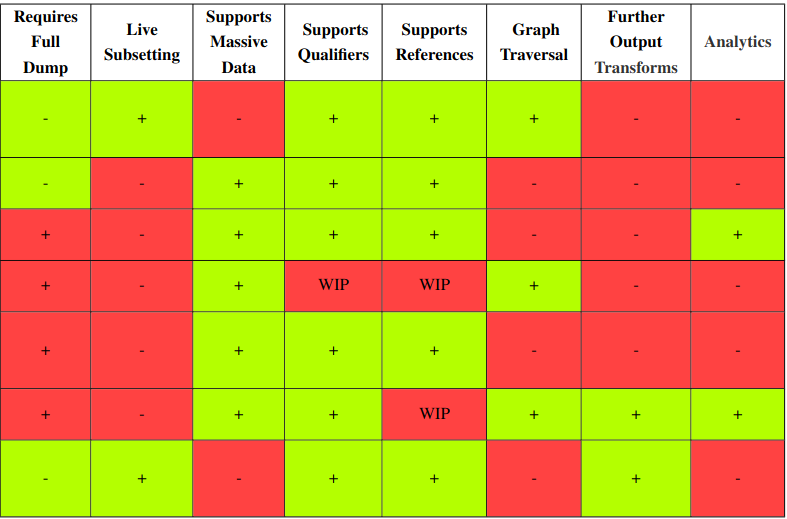
New paper: "Wikidata subsetting: approaches, tools, and evaluation"
Just before the end of the year, the Wikidata subsetting: approaches, tools, and evaluation paper by Seyed Amir Hosseini Beghaeiraveri et al. got published (doi:10.3233/SW-233491). I am really excited our group (i.e. Ammar and Denise) has been able to contribute to this. I think it also is a great example of the power of hackathons to bring together people. -
PhD Defences: Andra Waagmeester and Marvin Martens
2023 has been a long year in which a lot happens. Two EU projects ended (RiskGONE and NanoSolveIT; more about that in a later post), our group leader Chris Evelo will retire this year, the ELIXIR Toxicology Community started (see this post), the new WikiPathways website launched (see this post), and a lot, lot more. -
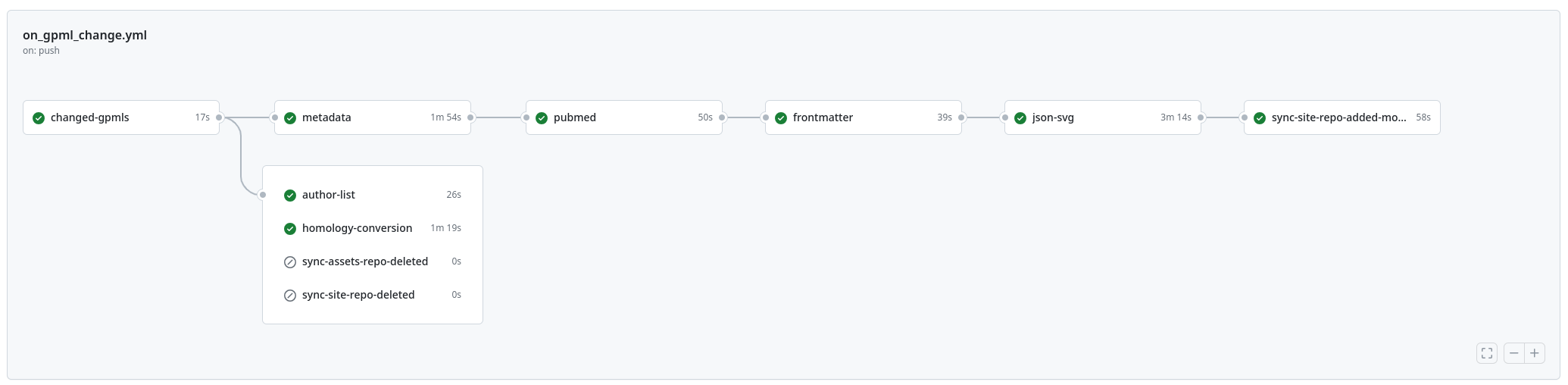
New paper: "WikiPathways 2024: next generation pathway database"
This week the next WikiPathways NAR Database issue paper was published (doi:10.1093/nar/gkad960). It is the next paper in a series of papers about the evolution of the Open Science project for making biological pathways available in a Open and FAIR way. This year, it described that significant move away from MediaWiki. It simply was too costly to keep up with the upstream code base (think: more than 200 thousand euro costly). This paper describes a transition to a modular system with Jekyll and Markdown as new platform technologies. The full details are available as open notebook science: everything is basically a git repository. -
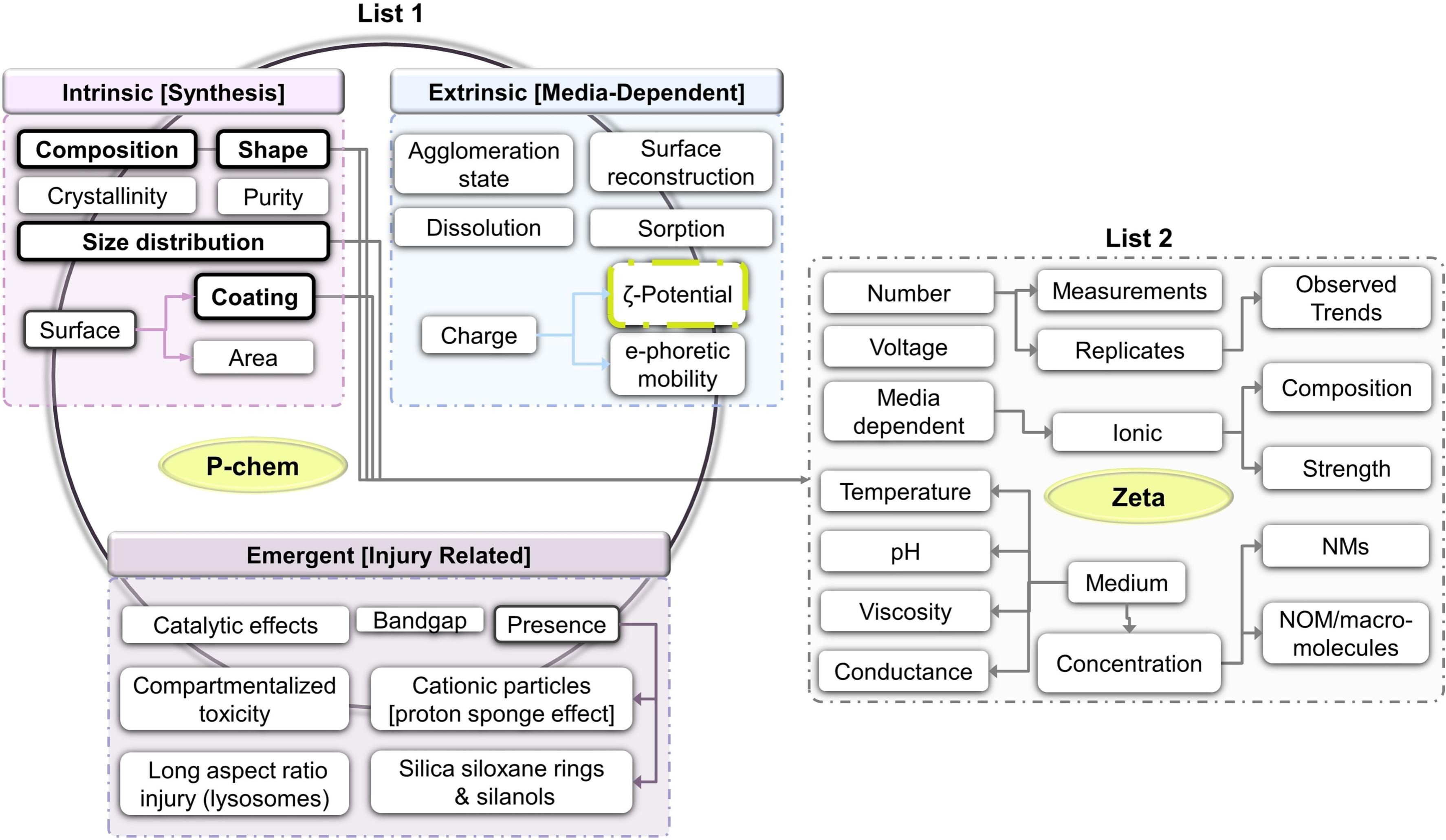
Using FAIR to select data for reuse
This paper got published in July already, but I had not had the time yet to blog about this exciting work by Irini Furxhi and Ammar Ammar: A data reusability assessment in the nanosafety domain based on the NSDRA framework followed by an exploratory quantitative structure activity relationships (QSAR) modeling targeting cellular viability (doi:10.1016/j.impact.2023.100475) -
Making BridgeDb Derby files with Groovy
I just want to drop this here. There are various ways to make BridgeDb identifier mapping files. Some of the tools predate my joining the BiGCaT research group and the BridgeDb project, but this Groovy page is basically what we have been using to create the metabolite identifier mapping databases: -
American Chemical Society Fall 2023 meeting
About four weeks ago the Fall 2023 American Chemical Society meeting (#ACSFall2023). I have attended a few ACS meetings in person and even organized a symposium at the 2010 ACS meeting in Boston. This time too, I did not participate in person, tho visiting San Francisco again would have been nice. I gave two presentations (slides doi:10.5281/zenodo.8255394), but have not uploaded my slides of the first presentation to Zenodo yet.