-
Two meetings: ELIXIR Toxicology and FAIR4ChemNL
Noting that in the coming week I am not attending the ELIXIR All Hands in Uppsala. Having lived in (and around) Uppsala for more than three years, I am disappointed and with the first stories from colleagues coming in even more. But it has been a way too busy year, I have much to finish up, and I need to take care of myself too. I am not 32 anymore. -
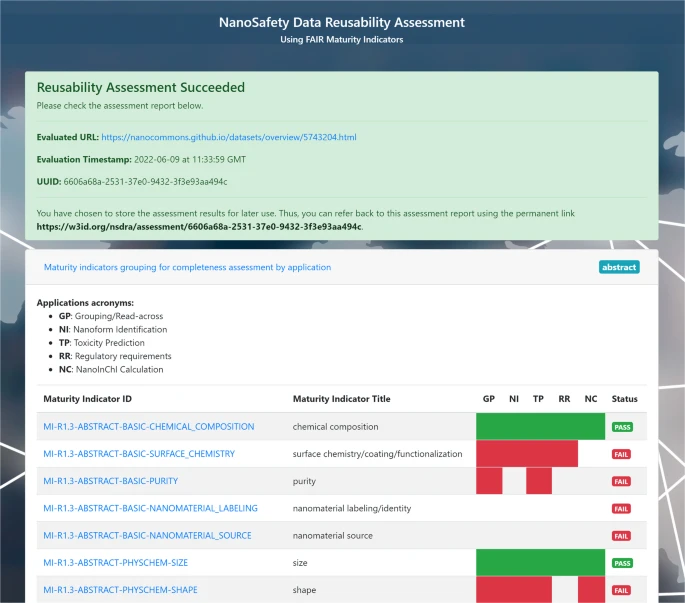
New paper: FAIR assessment of nanosafety data reusability with community standards
Ammar is finishing up his PhD thesis with his research on the use of FAIR towards predictive toxicology. Or, “AI ready”, as the term FAIR is now sometimes explained. Any computational method needs good data, and just FAIR is not enough. It needs to meet community standards, as formalized in R1.3. To me, this includes meeting community standards like minimal reporting standards. Indeed, in the EU NanoSafety Cluster the notion that FAIR data also needs be scientifically good data is well noted. -
New paper: A template wizard for the cocreation of machine-readable data-reporting to harmonize the evaluation of (nano)materials
I was about to call this blog post From spreadsheets to RDF, after the post last week. But then I decided to just use the pattern I typically use. Why I wanted to use that shorter term in the first place was that one of the thing I like about the AMBIT software (of OpenTox and eNanoMapper fame) is its RDF support (see doi:10.1186/1756-0500-4-487). But RDF, ontologies, those are hard things. And unlike mathematics, we do not have simple objects like integer numbers or simple operators. Well, I think we do, and we talk about them. But there is no obligatory education. Just like any biologist needs to know what 1 + 2 means, I think any biologist needs basic knowledge about how knowledge graphs work. But sometimes feels like a taboo, like cursing in the life sciences church. -

New paper: From papers to RDF-based integration of physicochemical data and adverse outcome pathways for nanomaterials
Making something FAIR is hard, particularly when you do more than making something findable. We’ve seen before that making something usefully findable requires deep indexing, and already that continues to be difficult, because we are not seeing it enough. So, when I thought convert a paper led by Hoet’s lab in Leuven into machine-actionable RDF to make it FAIR, I gravely underestimated the amount of work. Jeaphianne et al. did an awesome job on this work (doi:10.1186/s13321-024-00833-0). -
cdk2024 #2: publishing grant proposals
Publishing grant proposal is still not very common. The proposal published in Research Ideas and Outcomes) (doi:10.3897/rio.10.e124884) for the NWO Open Science grant for the CDK is, however, not the first and hopefully not the last. Interestingly, it is already cited in (the German) Wikipedia. It is used there to support a statement which tools use the Chemistry Development Kit. -
cdk2024 #1: NWO Open Science grant for the Chemistry Development Kit
We recently got awarded our second NWO Open Science grant (OSF23.2.097), this time for the Chemistry Development Kit (CDK). “We” here is me and Alyanne de Haan, René van der Ploeg, and Marc Teunis from Hogeschool Utrecht. The proposal has been submitted for public dissemination in RIO Journal, like we did with the first NWO Open Science grant. -
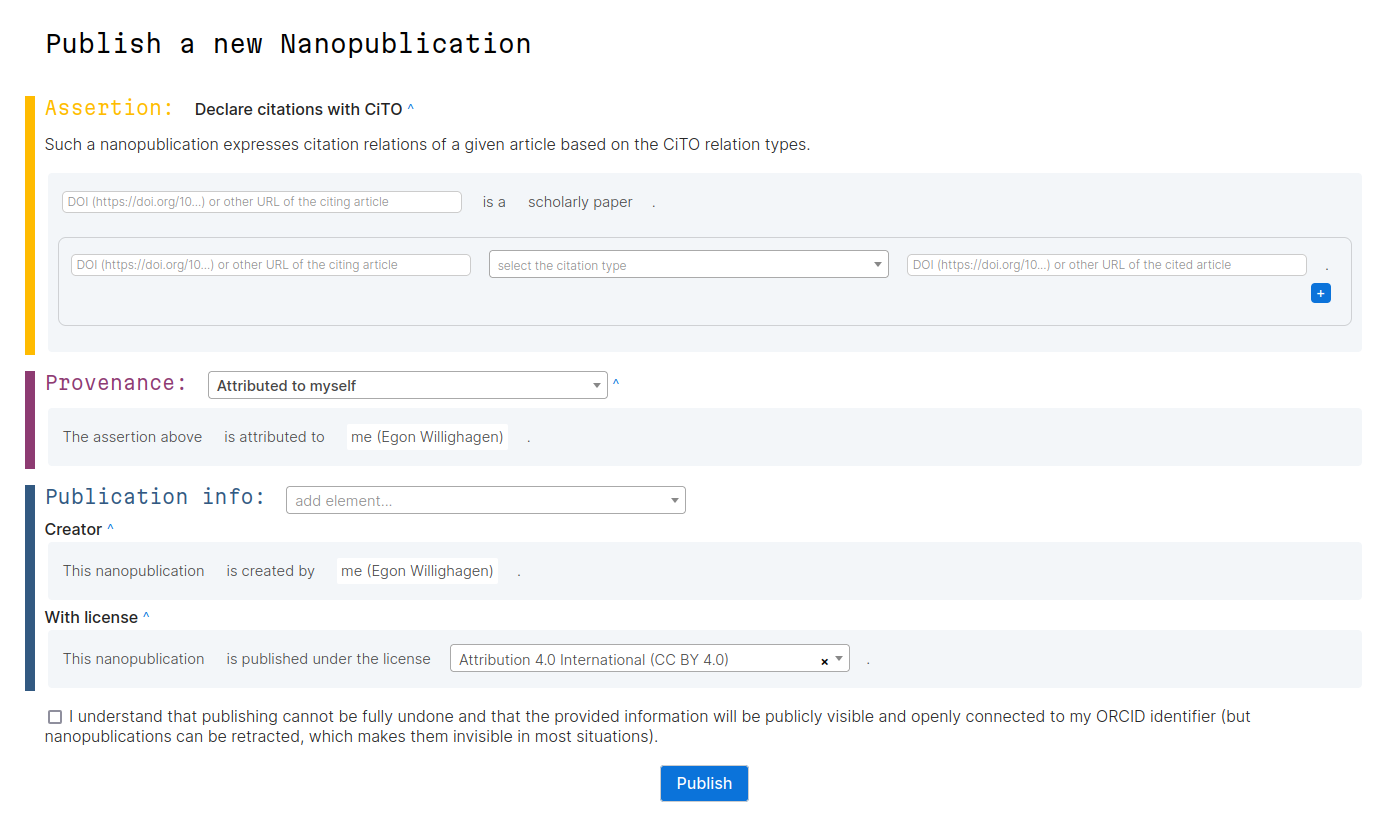
Open Science Retreat #2: CiTO Nanopublications
During the Open Science Retreat I organized a short session where we looking into typing citation intentions using a new nanopublication template. First, let’s describe nanopublications (originally used in doi:10.3233/ISU-2010-0613) a bit. Scholia gives a nice overview of (macro?)publications on the topic. The nanopub.net website describes that [a nanopublication is a small knowledge graph snippet with metadata that is treated as an independent (scientific) publication.]. The knowledge graph, it continues, can be anything from an opinion to the link between a disease and a gene (doi:10.1109/ESCIENCE.2018.00024).