-
Where's the maize genome torrent?!?
/. just posted a story about the maize genome just published, for which the sequences can be downloaded from this FTP site. The files are not that large at all. But it makes me wonder… where are the .torrent files for the sequenced genomes? Here’s Davids catch on the story. -
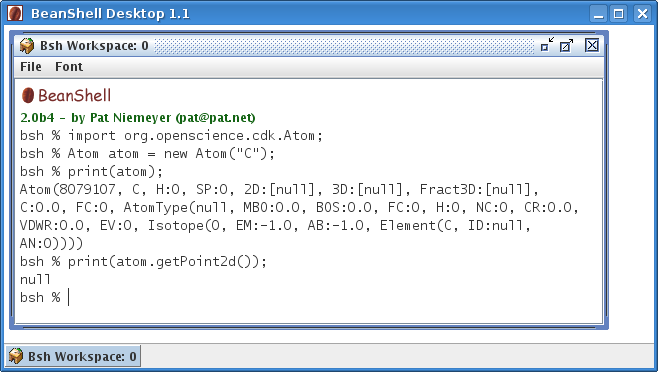
CDK is now available from your nearest Debian mirror
Some days have passed, and the Debian mirrors have now picked up the CDK package (unstable only so far), allowing you to sudo aptitude install libcdk-java from your favorite local mirror. The details are available from this packages.debian.org/libcdk-java page. The fact that it is listed as contrib is a small mistake; the package is really main material. -
CDK close to entering Debian
Michael Koch (aka man-di) and Daniel Leidert (as part of the pkg-java team) have worked on packaging the CDK. The ran into some issues, such as the CDK build system not perfectly compatible with the Debian java libraries in /usr/share/java. Both detection of the available libraries as well as putting them in the classpath, caused trouble with the CDBS-based build system wrapping around the Ant build.xml (note the many commit this weekend ;). -
Simple, Open Bug Track System: social bookmarking
Jim replied to the request by Anthony in my blog for a bug track system for CrystalEye (in beta), after a discussion on the CIF processing pipeline (see here, here, here and here). -
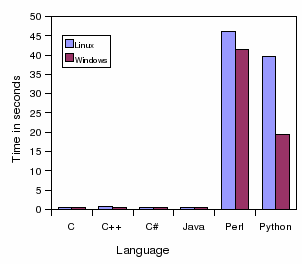
Performance: C, C++, C#, Java, Perl and Python
Mathieu Fourment (et al.) just published a paper on some performance testing on 6 programming languages in BMC Bioinformatics: A comparison of common programming languages used in bioinformatics (doi:10.1186/1471-2105-9-82). The below figure is from the paper, for a sequence alignment exercise (copyright with paper authors, OpenAccess license of journal): -
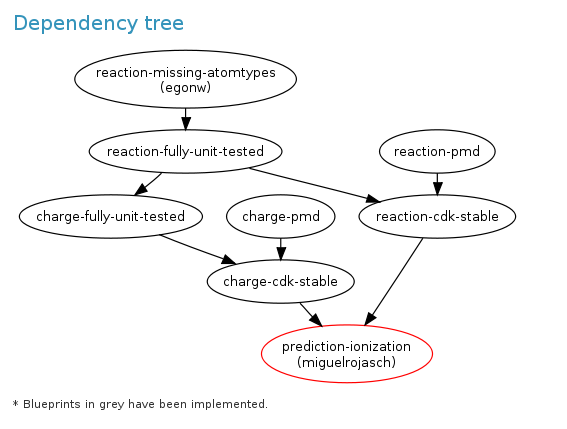
Defining Development Goals: LaunchPad complements SourceForge
Today, Miguel (who made the 10000th CDK commit) and I gave LaunchPad a go, because if offers a nice GUI for planning and monitoring source code development. We have set up a CDK team and a CDK project. LaunchPad has overlap with SourceForge functionality, but they idea is not to duplicate functionality. Moreover, we do not translate the CDK either, so that LaunchPad functionality is not useful either. Not for the CDK at least; maybe for Jmol and Bioclipse?
