-
Adding a new dictionary to Oscar
Say, you have your own dictionary of chemical compounds. For example, like your company’s list of yet-unpublished internal research codes. Still, you want to index your local listserv to make it easier for your employees to search for particular chemistry you are working on and perhaps related to something done at other company sites. This is what Oscar is for. -
Oscar4 command line utilities
One goal of my three month project is to take Oscar4 to the community. We want to get it used more, and we need a larger development community. Oscar4 and the related technologies do a good, sometimes excellent, job, but have to be maintained, just like any other piece of code. To make using it easier, we are developing new APIs, as well as two user-oriented applications: a Taverna 2 plugin , and command line utilities. The Oscar4 Java API has slightly evolved in the last three weeks, removing some complexity. In this post, I will introduce the command line utilities. -
CiteULike CiTO Use Case #1: Wordles
Last month I reported a few things I missed in CiteULike. One of them was support for CiTO (see doi:10.1186/2041-1480-1-S1-S6), a great Citation Typing Ontology. -
Oscar4 Java API: chemical name dictionaries
Besides getting Oscar used by ChEBI (hopefully via Taverna ), my main task in my three month Oscar project is to refactor things to make it more modular, and remove some features no longer needed (e.g. an automatically created workspace environment). Clearly, I need to define a lot of new unit tests to ensure my assumptions on how to code works are valid. -
Cb: New Blogs #14
Just a few new blogs since #13 in July : -
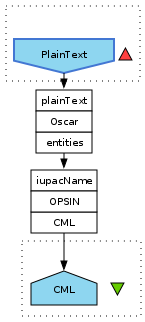
Oscar text mining in Taverna
One of the goals of my project in Cambridge is to make Oscar available as Taverna plugin (source code, Hudson build). I have progressed somewhat, but still struggling with getting the update site working. The plugin actually installs into Taverna 2.2.0, but the activities do not show up. While this is work in progress, and the other project goal is refactoring, a current demo workflow looks like: