-
Cb: New Blogs #13
The Cb software is still holding… I jettinsoned the old post cache, which speeded up the processing of blogs considerably, but the system just doesn’t scale right. Yet, Euan has done a great job, and the Cb site has now been online for some three years! Here are some new blogs included in the aggregation and analysis: -
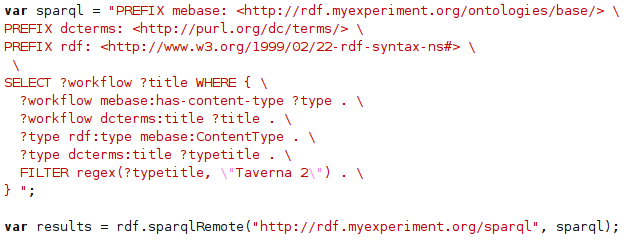
ChemPedia RDF #1: the SPARQL end point
Well, you might spot a pattern here; yes, another chemical SPARQL end point (actually, it shares the end point with the Solubility data). This time around Rich’s ChemPedia. Taking advantage of the CC0-licensed downloads , I have created a small Groovy script (using this JSON library) to convert the ChemPedia JSON into Notation3: -
Really free chemistry books
With pleasure I read Analogue or Digital? - Both, Please. Funnily, I just created MP3 (or, preferably Ogg Vorbis, superior but hardly any support by commercial companies, who rather seem to pay license fees) directly from the CD. -
The Social Web does not wait for Bioclipse... here comes Google Wave
Google Wave is going to change the web. It’s the end of Google Docs, and likely many other services. It’s going to be Open Source and being a Wave Provider will not be restricted to Google. This will be enough to make this a success. If you haven’t watched the full video demo yet, please have a look yourself: