-
Groovy Cheminformatics 4th edition
Six month was not quite the amount of time I anticipated between the third and fourth edition, but I finally managed to upload edition 1.4.7-0 of my Groovy Cheminformatics book. The first three editions sold 37 copies, including two for myself. Enough to feel supported and to continue working on it. -
ChEMBL-RDF: Uploading data to Kasabi with pytassium
I reported earlier how to I uploaded the ChemPedia (RIP) data onto Kasabi. But for ChEMBL-RDF I have used the pytassium tool, not just because it has a cool name :) I discovered yesterday, however, that I did not write down in this lab notebook, what steps I needed to take to reproduce it. And I just wanted to uploaded new triples to the ChEMBL-RDF data set on Kasabi. -
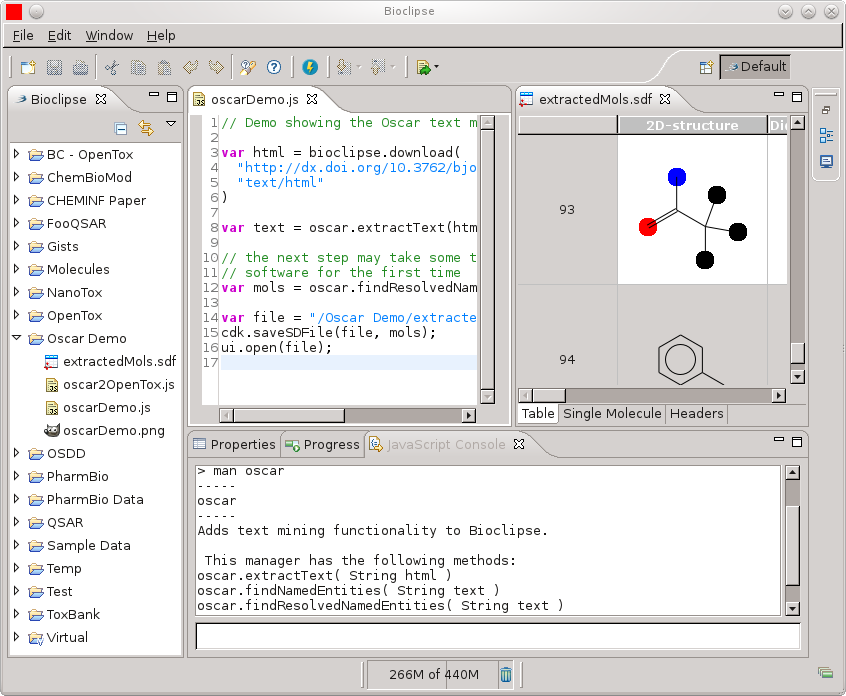
Bioclipse-Oscar4 - Text mining in Bioclipse
Almost a year ago I started a position with Peter Murray-Rust to work on Oscar for three months (see this overview of results; a paper by the full Oscar team (Sam, David, Dan, Lezan) is pending, and I’m really happy to have been able to contribute bits to the project). Since then, I have had little time :( That’s how it goes, with post-hopping, unfortunately. One thing I did do after that, was write a Bioclipse plugin. -
InChIKey collision: the DIY copy/pastables
About two weeks ago, the ChemConnector blog reported an InChIKey collosion detected by Prof. Goodman . Unlike the previous collision, this one was based solely on the graph and not on stereochemistry. The two molecules both have the InChIKey OCPAUTFLLNMYSX-UHFFFAOYSA-N: -
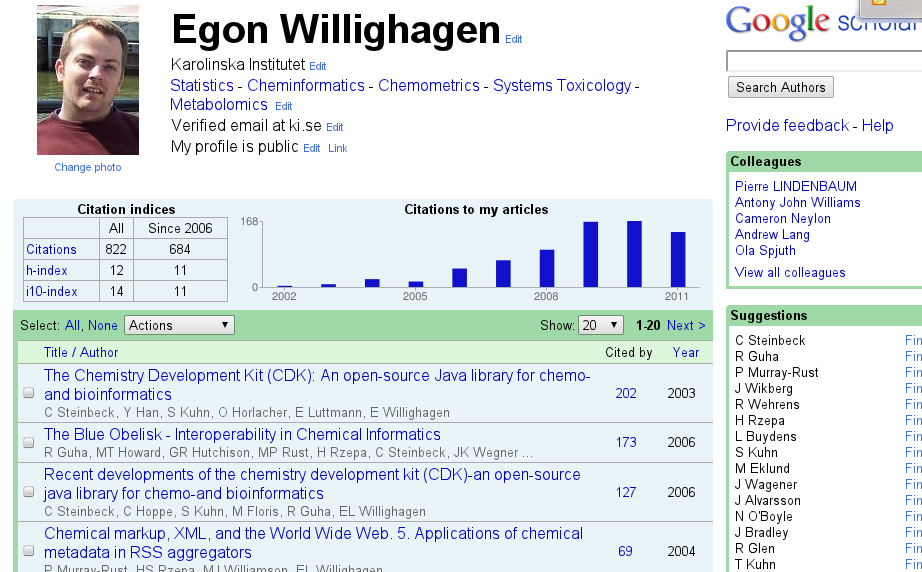
My Google Scholar Citations profile arrived
Web of Science is my de facto standard for citation statistics (I need these for VR grant applications), and defines the lower limit of citations (it is pretty clean, but I do have to ping them now and then to fix something). The public front-end of it is Researcher ID. There is an Microsoft initiative, which looks clean but doesn’t work on Linux for the nicer things, but the coverage of journals is pretty bad in my field, giving a biased (downwards) H-index. And CiteULike and Mendeley focus more on your publications than on citations (though the former has great CiTO support!). -
Groovy Cheminformatics 3rd edition
Update: the fourth edition is out.
