Bioclipse-Oscar4 - Text mining in Bioclipse
Almost a year ago I started a position with Peter Murray-Rust to work on Oscar for three months (see this overview of results; a paper by the full Oscar team (Sam, David, Dan, Lezan) is pending, and I’m really happy to have been able to contribute bits to the project). Since then, I have had little time :( That’s how it goes, with post-hopping, unfortunately. One thing I did do after that, was write a Bioclipse plugin.
I was asked recently via LinkedIn if I was planning a Bioclipse-Oscar plugin, and
I realized that I forgot to blog about it. So, here goes. The oscar manager I implemented follows the
Oscar API , and these
methods are available: extractText(), findNamedEntities(), findResolvedNamedEntities().
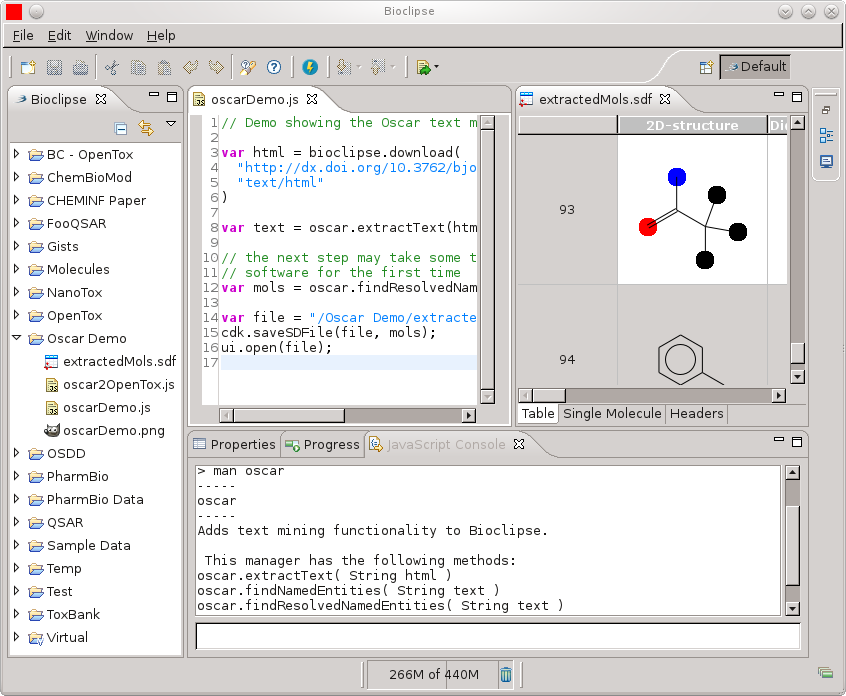
When I wrote the plugin, I also uploaded an example workflow to MyExperiment. The code is:
// Demo showing the Oscar text mining functionality
// in Bioclipse
var html = bioclipse.download(
"http://dx.doi.org/10.3762/bjoc.6.133",
"text/html"
)
var text = oscar.extractText(html);
// the next step may take some time, while
// initializing the Oscar software for the
// first time
var mols = oscar.findResolvedNamedEntities(text);
var file = "/Oscar Demo/extractedMols.sdf";
cdk.saveSDFile(file, mols);
ui.open(file);
The code will extract chemical entities, and open a molecules table in Bioclipse: