-
INTOXICOM Workshop #4: Training Materials for Toxicology
INTOXICOM is an ELIXIR Toxicoloy Community (doi:10.12688/f1000research.74502.2) implementation study, organizing five workshops, each one with a different topic. The first three workshops were in Utrecht (doi:10.37044/osf.io/un2rw), Basel, and Uppsala. The fourth workshop has the theme training material and will be held in Birmingham (thanks Iseult!). And one aspect of that is to make existing training material from toxicology projects indexed and therefore more FAIR. -
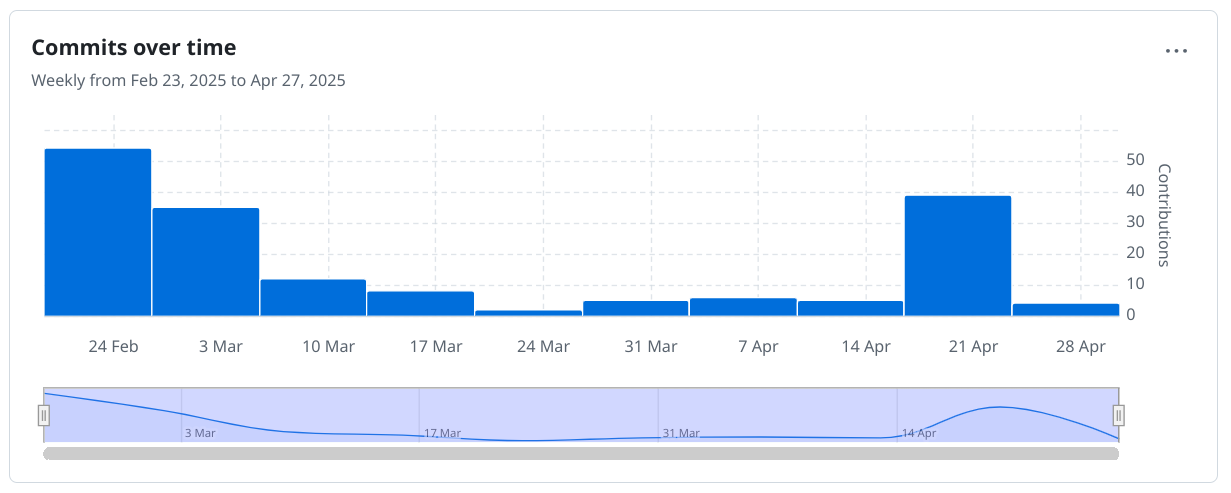
The April 2025 Scholia hackathon
This is the third weekend I am working on Scholia, the first two part of the April 2025 hackathon. It follows the hackathons last year October and November hackathons. There is some urgency for this unpaid work, because Wikidata is splitting the RDF into two SPARQL endpoints (see this The Signpost and this post by Finn). This split has happened, but there is a legacy server for tools that have not been upgraded. -
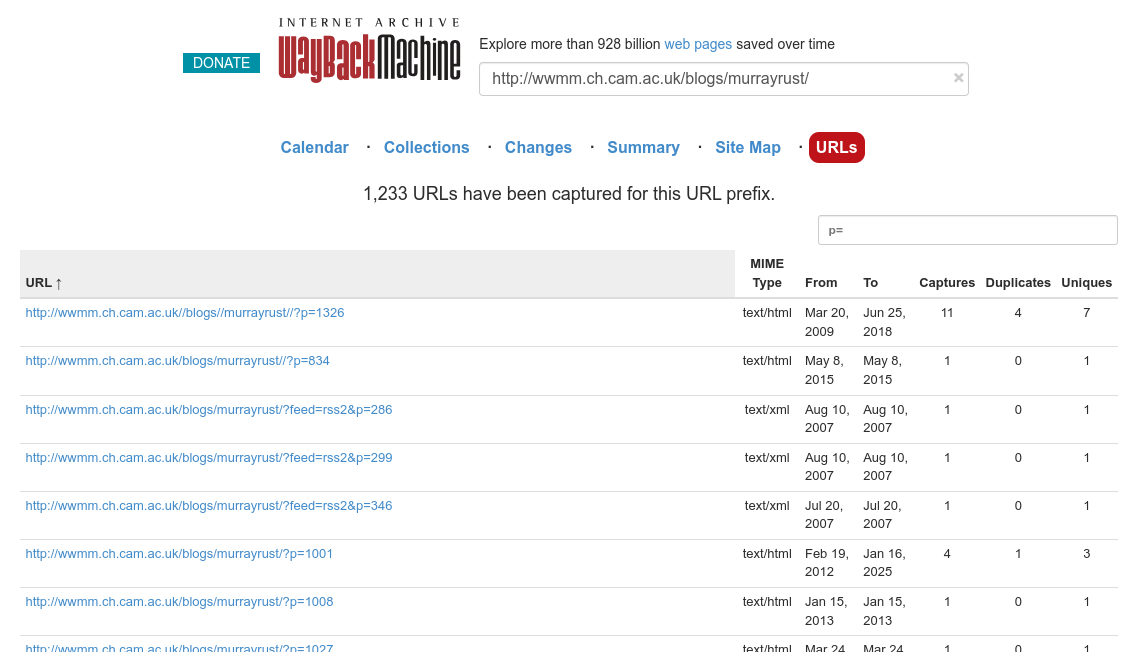
Updating links to blogs: Peter Murray-Rust
One reason to move my blog from Blogger to a git-backed repository is that I can update links but that the version history shows exactly what change was made. I have been using three icons: when I can find a new URL for the website, I use ; when I cannot find a new URL, but the old URL is in the Internet Archive, I use a ; finally, if I cannot find anything to replace the broken link, I use . For example, the blog of Rich Apodaca is now archived, and I have been updating the many links to his (still running) blog to use DOI links. That has the added benefit of making blog-to-blog citations more FAIR. -
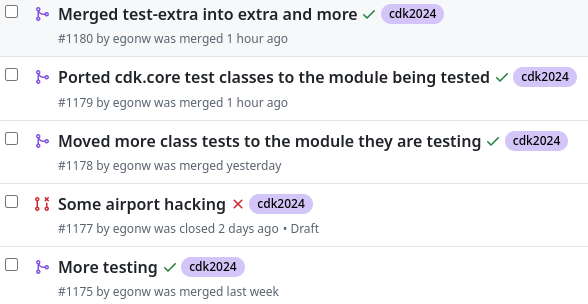
cdk2024 #6: wrapping up already
Tomorrow is already the last day of the NWO Open Science grant for the Chemistry Development Kit. We are wrapping up, but I am happy we have a few weeks more to finish up the reporting. We held a user group meeting earlier this month (btw, check out the slides by Jonas), and I did a few more JUnit testing updates last week: -
New preprint: "BioHackEU24 report: ORCID and ROR identifiers in BioHackrXiv reports"
While this was not the primary hack project during the ELIXIR BioHackathon Europe last autumn, but I really like BioHackrXiv and I got the question if I could have a look at getting the ORCID logo in generated PDF. The ORCID was already in the YAML metadata of report markdown, so it sounded easy. Well, it was a big more complicated, but all the nicer to now have the project report online (doi:10.37044/osf.io/p9u42_v1). And once the ORCID was working, adding the Research Organisation Registry ID was not much harder. Cool to see both used in other recent BioHackrXiv reports! -
cdk2024 #5: Chemistry Development Kit User Group Meeting - Day 2
Where the first workshop day had several talks about new and old features of the Chemistry Development Kit (CDK), the second day was a hackathon day. We hacked and we talked. The coding was not mostly only the CDK repository itself, but some things happened there too: