One Million IUPAC names #2: the 100 thousand milestone
Two and a half month into the One Million IUPAC Names project, we passed the third milestone, the one for 100 thousand IUPAC names (doi:10.5281/zenodo.15266459). Time for an update.
This milestone release took a bit longer. Going from 50 to 100 thousand is a bigger step than from 10 to 50 thousand, but the open access chemistry literature was already done by then. Basically, I ran out of open access chemistry publications. The scripts are now finding names in all (open access) literature, and the number of new names per articles is a lot lower. Still about 1 in every twenty to 30 articles. But the diversity in names is not really going down, which is important.
The first few weeks, I used the Google Colab to run a Jupyter notebook, initial created by Magnus, but having to process more articles to get a reasonable number of new IUPAC names required longer and longer jobs, and then Google Colab is not really fit (well, the free version anyway). So, I started using a local script. That turned out to be able to handle up to 20 thousand articles in one go and runs at least twice as fast. Moreover, I can run three of them in parallel.
And that had impact. With each commit around 1000 new IUPAC names, the number of commits went up remarkably last week:
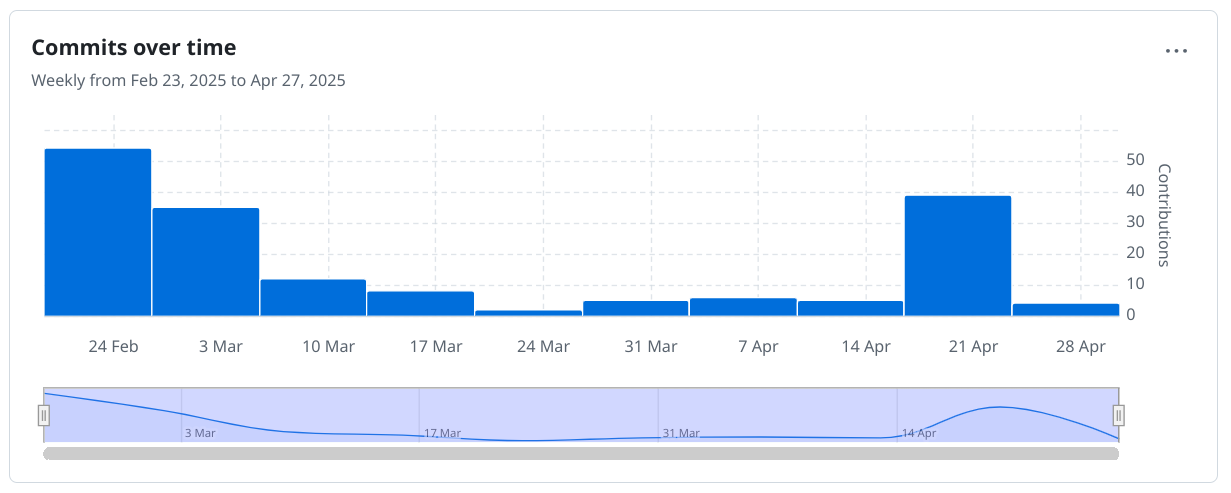
At the current speed, I think we’ll make it to 150k soon and I added a new milestone for 200k, which sounds doable in the next three week. That also means that 1M extracted IUPAC names from literature has become a reasonable goal. And we can start thinking about the 2, 5, 10, 50 and 100 million IUPAC names. Those are, at the current speed, rather unlikely to reach from the open access literature anytime soon. That brings us to the question, what will. Well, I have some ideas.
Idea 1: name variations
First, I am figuring out some ways to make variants of names (no, not based on hyphens and spaces; that’s too easy), but actual variations of the chemical structures. For example, I could exhaustively replace “methoxy” with “ethoxy”, and iterate the halogens and acyl chain lengts. I have little doubt that I can grow the list with this approach easily a 5-fold, maybe even a 10-fold.
Idea 2: hallucination
Another idea is that I could use tools that can generate IUPAC names for a limited set of compounds. I once wrote code for alkanes myself and if I can find that, I may be able to generate additional names. But perhaps more realistic is that I train a deep learning model and have it generate names for all compounds in Wikidata (~1.5 million) or PubChem (>100 million). STOUT needed 81 million compounds (doi:10.1186/s13321-021-00512-4), but I don’t need a good model; I just need a model that comes up with new, valid names. Hallucinated names, but valid.
While the list of valid names grows, I can retrain the deep-learned model and repeat. As long as the diversity remains high enough, one could hypothesize that the deep learning will learn new tricks. And then, that should be a near infinite source of additional names.
Idea 3: (semi-)closed access literature
Also, I haven’t touched closed access articles yet. This is all based on the collection of full texts in Europe PMC. For example, I could start with the green open access article in (Dutch) university repositories, particularly those with large chemistry departments. PDF to text tools are mature enough that this will provide a new source. Oh, and perhaps PhD thesis, which are now also increasingly archived in university repository under open access. And that reminds me of a Dutch project two decades ago doing exactly that. I wish I remembered the name.
Idea 4: alternatives to Oscar4 and Europe PMC
So, the first round of named entity recognition was with Europe PMC itself, as explained in the first post. The move to Oscar4 helped a lot. But there exist many other chemical NER tools, like (doi:10.1093/bioinformatics/btn181. And those may find an additional number of names, even with just the literature I already covered.
Well, you get the idea.
ICCS poster rejected
Unfortunately, the ICCS poster abstract did not make the cut. The score was high enough, but they received many abstracts and had to make a selection (of course, I am part of the ICCS organization, and have more details of how it came about). I really like the project, and eager to write up a paper around it.