Where does the WikiPathways Cited In information come from?
I have been wanting to blog about this since this summer, but with everything going on, I never really got around to it. What is this Cited In feature of WikiPathways and where does that information come from? If you have not noticed this yet, this is what it looks like for WP4846:

Recently, I was close to writing up the context, because it is related to a new feature of the profile pages, where you now can look up citations to pathways that you first authored (see this post). And it also relates to the data I have been collecting around citation intention annotations: articles that cite one of the WikiPathways papers and mention a specific pathway, could be considered cito:usesDataFrom (see doi:10.1186/s13321-023-00683-2).
A third angle to citations to specific WikiPathways is the following. WikiPathways is used a lot in data analyses and putting experimental data in biological context. How researchers do this varies a lot, in multiple ways. But just thinking about this factually, research output cite specific biological pathways. And there are some interesting phenomena there. Back in 2015 at the Metabolomics Society meeting in San Francisco (apparently, I only blogged about the meeting only once?), when I visited the 500+ posters looking for interesting biological pathways, there were a lot of studies on different species, different diseases, different toxicities. The biological response had one thing in common: it always was the TCA cycle that was key (see doi:10.1096/FJ.11-203091 for a 2012 comparison of TCA models).
Thus, with so many articles mentioned specific pathways and deriving biological knowledge from this, what is reasonable to expect? Do we expect co-citation effects? That is, if two articles found the same set of pathways of interest to their data, is the data showing a similar biological response? Do we expect a similar thing like the above TCA cycle in metabolomics, something similar to the notion of frequent hitters (see doi:10.1021/jm010934d)?
Of course, to test this hypothesis we need data and the Cited In feature comes in. At the time of writing of this blog post, we can see on this page that 878 pathways have been cited a total of 2715 times. We are getting somewhere. This blog post will not analyze this data, which is one reason why I had not blogged about it. But from the above you can understand that I want to :)
The Cited In feature
This Cited In feature was introduced along with the new website (see doi:10.1093/nar/gkad960), where we change how GPML files are stored and how web pages are created from that. Because we are no longer confined to the MediaWiki platform (which has served the project for very long, very effectively), it is easier to integrate information from other sources. For example, from literature databases. This feature was developed by Alex Pico at the Gladstone Institutes (see this 2022 commit), where he uses the NCBI eUtils API to access PubMed Central. The data is then collected into this YAML file which then gets used to generate webpage content (like the section in the above screenshot and the page mentioning the current statistics).
Where is the data coming from?
As just explained, originally the data was only coming from NCBI. However, because I found many articles citting specific pathways that were not picked up by this approach, and I wanted more data, so I started searching Europe PMC the European partner of PubMed Central. However, I am not automating this. I want to see the data, the articles, and how people cite the pathways. I need to see that so that I can better understand how people are using the data/knowledge from WikiPathways. I cannot keep up with checking why people are citing my own research, but I once was. I learn(-ed) a lot from that.
I normally use a search that requires the word “WikiPathways” to be
mentioned in the article (in most, but
not all of them; citing literature you extend sounds like a core scholarly value, but is factually
not systematically complied with), and then manually searching for “WP”. With close to 1000
PubMed Central articles mentioning WikiPathways in 2025 and that these are mostly full texts,
I can see if the cite specific pathways. A good number of article mentions the WikiPathways
identifier, e.g. the aforementioned WP4846. If the article only mentions a pathway title,
I cannot confidently identify which pathway is cited, so I exclude that.
I originally started out manually editing the YAML file where the citations are collected, but by now use a script similar to Alex’ R script. This makes it far easier to scale up, as I just have to populate a three column TSV file, which is used by my R script to update the YAML file. This manual approach ensures that I am not looking at text mining results, but see the citation of the WikiPathways identifier with my own eyes. That’s just how I like it.
The full history of the YAML file content can be found on this GitHub page and this git blame tells you if the information came from PubMed Central via the API, or was added by me:
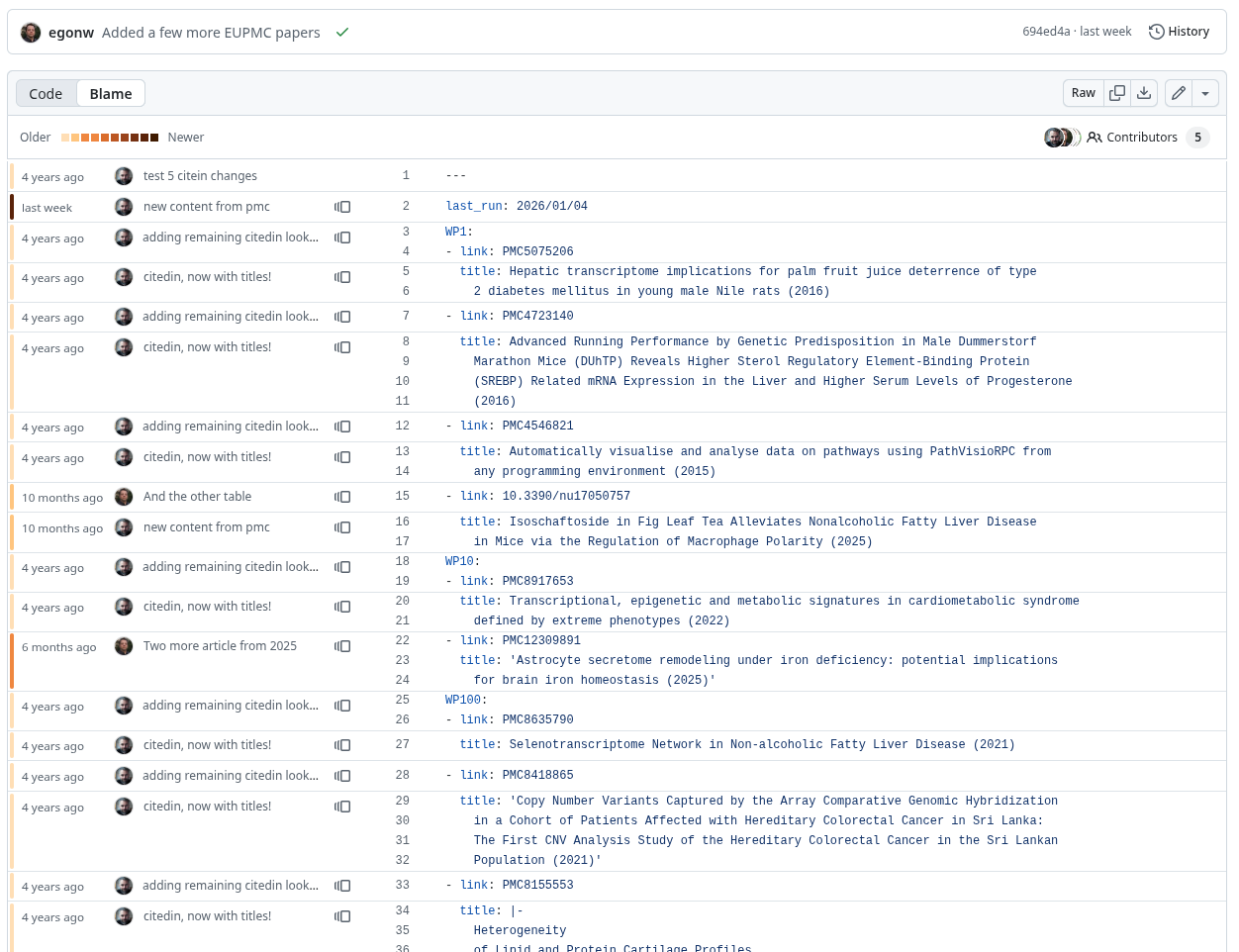
This is Open Science in action: added transparency and making it easier for anyone to verify, so that no one needs to be stuck in (dis)trust.
Of course, as we know from the CiTO ontology and real-world data, there are so many different reasons why journal articles are cited (just an example), the data in the YAML file and on the WikiPathways website in the Cited In feature does not have direct meaning. Just like a high citation count for an article or even a journal impact factor cannot be directly interpreted (despite so many researchers just blindly doing just that).
What’s next?
Well, while I did not do any analysis yet, and do not even know yet how much citations we need to reach some level of statistical significance, there are some observations I can mention:
- if your analysis included anything like linking your data to pathways, citing those pathways is a good way to give credit to the researchers that created that pathway
- if you cite data, please cite that as accurately as possible, see e.g. DataCite
- I wish all journal articles citing specific pathways from WikiPathways would include the pathway identifier
- I congratulate those authors that even mentioned the revision of the pathway! well done!
And about biological interpretation, our group has long published that some genes with differential data mapping to a pathway does not imply that that pathway is really affected. Gene-set enrichment and over-representation analysis are a starting point; not a conclusion. I wish more people were more aware of the work in our (now) Translational Genomics research group. Like that of Martina Kutmon (now as MaCSBio2), whom I have had the pleasure of collaborating with for quite some years now (and long time archtect of WikiPathways).
There is so much more I want to write up about WikiPathways, but I leave it to this for now.
You can use your Mastodon or other ActivityPub-based Fediverse-account to comment on this article by leaving a publicly visible reply to this associated post. Content warnings are supported. You can also delete your comments at any time by deleting your post on the Fediverse.