CiTO updates: Wakefield and WikiPathways
This summer I am trying to finish up some smaller projects that I did not have time for to finish, with mixed successes. I am combing this with a nice Dutch staycation, and I already cycled in Overijssel and in south-west Friesland and learning about their histories. But this post is about an update on my Citation Typing Ontology use cases. And I have to say, a mention by Silvio Peroni is pretty awesome, thanks!
First, the bad news. I still did not get around to the following to tasks I have. First, I need to write up a step-by-step guide how to create CiTO nanopublications and matching draft article. Second, I still need to work out how to update the JATS workflow for CiTO annotation in BioHackrXiv.
Wakefield
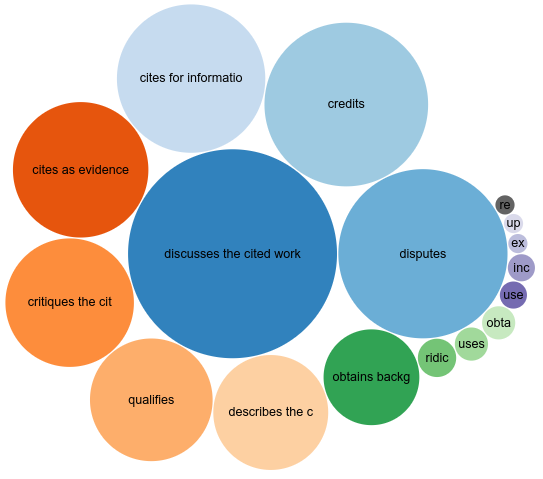
Let’s first start with a dataset. Peroni mentioned a study they did (10.1007/S11192-021-04097-5) into why the famous Wakefield paper (doi:10.1016/S0140-6736(97)11096-0) is cited. They published their data set on Zenodo (doi:10.5281/zenodo.13166142) with CCZero, so I imported it into Wikidata. Well, at least the citations of articles already in Wikidata. I used a Bacting (doi:10.21105/joss.02558) script and it actually was quite short. In the end, this added some 500 new citation intentions to Wikidata, now at almost 2000. This is also the third dataset with explicit CiTO intention annotations (see also this post).
This is what the CiTO section of the Wakefield paper in Scholia (doi:10.1007/978-3-319-70407-4_36) now looks like:
WikiPathways
A second thing I want to show is a potentional CiTO intention annotation dataset. Almost two years ago Alex Pico started a new WikiPathways feature as part of the new website (doi:10.1093/NAR/GKAD960)): a list of citations to specific pathways (in WikiPathways). Alex’ setup is fully automated and using PubMed Central and find mentions in figure captions:
Beyond citations to previous WikiPathways journal articles, we have identified 1228 mentions of a total of 582 unique WikiPathways pathway model identifiers, e.g. WP4846, in PubMedCentral articles over the past 13 years.
The file format is a pretty basic YAML file:
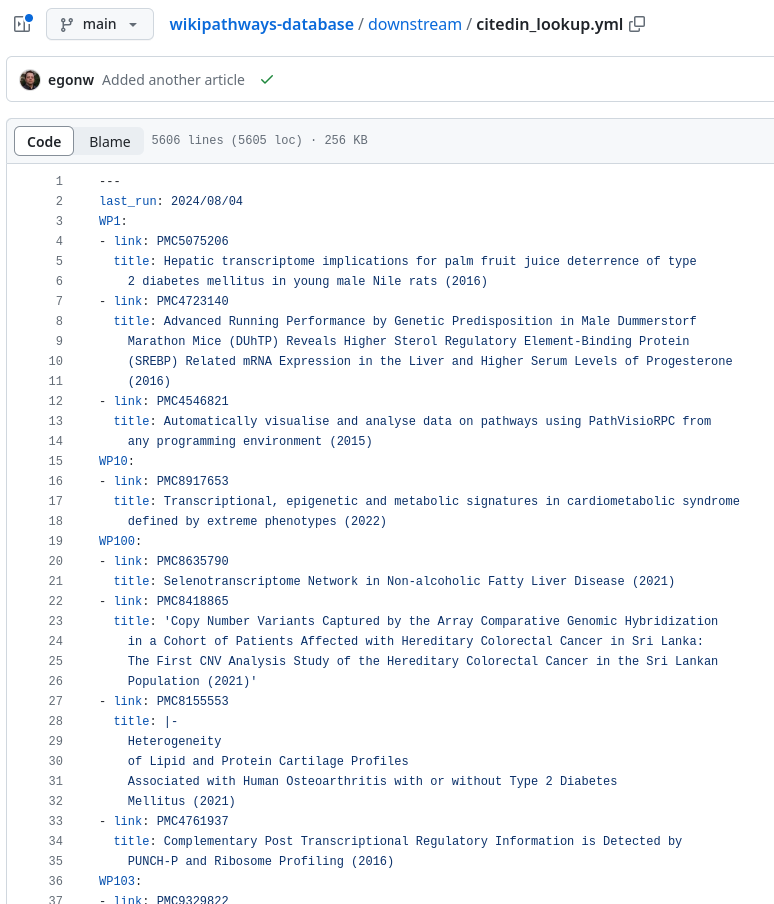
Additional mentions are found in the main text and tables in the article. These are not always picked up. These can be added manually. Over the past months and the past two weeks particularly, I have been adding additional mentions, not listed yet. We now passed 1500 mentions but I cannot easily give the other statistics.
BTW, anyone can add these citations with the ‘edit’ pencil and some Microsoft GitHub editing (but as far as I am concerned, please feel free to also just mention the paper on the WikiPathways Community Forum):

So, in the next few days I plan to do two things: 1. generate RDF for the YAML file and make that part of the
monthly WikiPathways RDF release; 2. extract citations and
offer this back to the OpenCitations project; and, 3. add the citations
into Wikidata. Of course, all with cito:usesDataFrom :)
There is a fourth things that I am still thinking about. I can also use the above data the annotation
citations to the WikiPathways papers if they also mention a WikiPathways identifier as cito:usesDataFrom,
but I cannot fully oversee the implications of that. What do you think?
You can use your Mastodon or other ActivityPub-based Fediverse-account to comment on this article by leaving a publicly visible reply to this associated post. Content warnings are supported. You can also delete your comments at any time by deleting your post on the Fediverse.