QSAR plugin for Bioclipse getting in shape
Over the last few weeks I continued the work on getting (descriptor-based) QSAR/QSPR implemented in Bioclipse. JOELib (GPL) and the CDK (LGPL) being two prominent opensource engines that can calculate molecular descriptors, and AMBIT a front-end.
To be able to do QSAR/QSPR model building from start to end in Bioclipse, I worked in April on an architecture for selecting descriptors . Being busy with so many things, it took me some time to get around to completing that, but here are the screenshots:
The funny characters and the whitespace is gone. Right now, it still only lists one provider, but I plan to add JOELib plugin soon. The list of actual descriptors is provided by the extension.
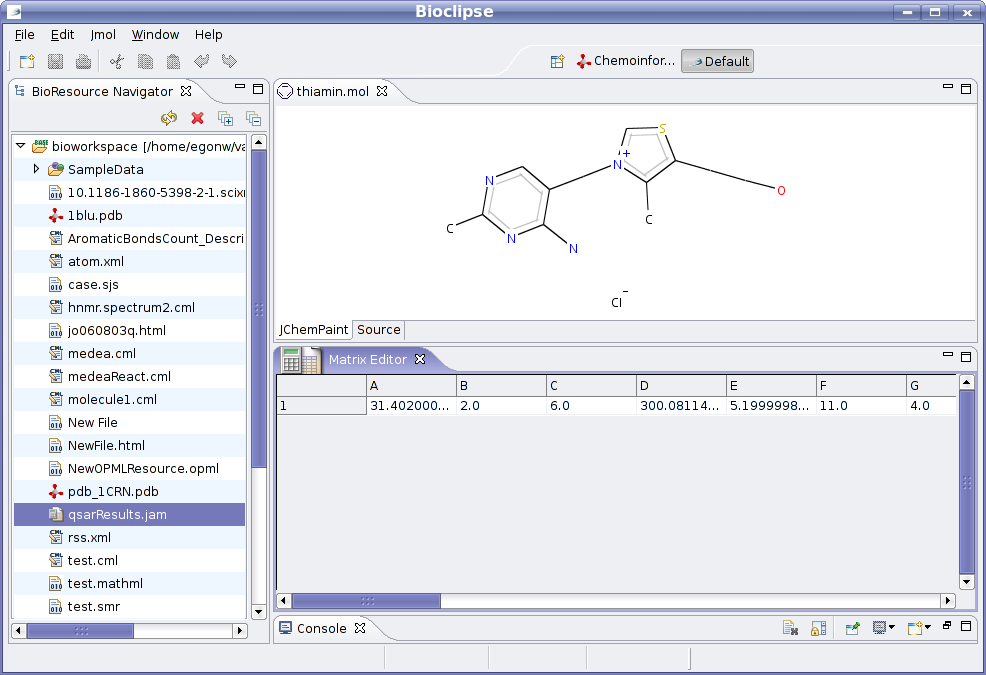
What Bioclipse then does, is have the extension calculate the descriptor values for the selected CDKResource in the BioNavigator
using the selected descriptors. This will then create a new MatrixResource in the Bioclipse workspace (currently called
qsarResult.jam), and which is opened in the Matrix editor:

There is still enough work left to do. For example, the columns are not yet labeled according to the descriptor name, and
selecting more than one CDKResource in the navigator does not give a multirow matrix yet.