Bioclipse now allows QSAR descriptor selection
In preparation for the Embrace Workshop for Bioclipse in May, I am working on the QSAR functionality of Bioclipse. A nice extension point got set up some time ago, called DescriptorProvider , and implemented by plugins to allow calculation of one or more descriptors for the selected molecules. Now, the functionality for the resulting matrix has been around for some time too.
What had not been available yet, was some GUI stuff to select descriptors to calculate, and the actual calculation. While the latter is yet to be hooked up, the selection of descriptors is now available:
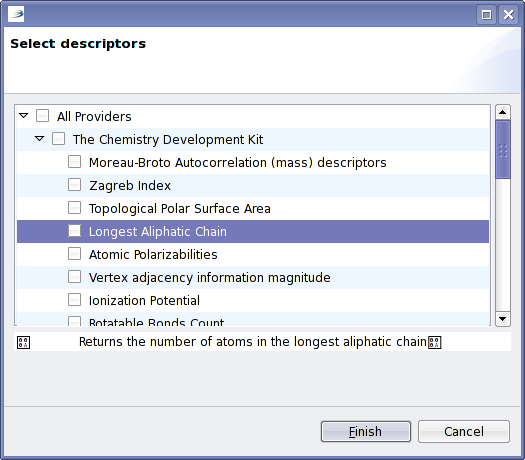
Interesting here is the use of OWL. CDK’s DescriptorEngine provides a simple API written by Rajarshi that interfaces to the dictionary support
for OWL (which CDK offers in addition to CML based dictionaries). All CDK descriptors are written up in OWL (the
source file
and the HTML version).
You’ll notice the weird characters in the screenshot; there something goes wrong with the encoding when reading the OWL.