Matrix support in Bioclipse
With chemometrics in mind (QSAR, data mining, …), I have started working on matrix support in
Bioclipse, because the matrix is the important step between (bio-)molecular content and statistical analysis.
I implemented this such that the actual matrix implementation can be freely chosen, that is,
bc_statistical provides a IMatrixImplementation extension point.
The plugin bc_jama provides a JAMA
based extension for this, but other implementations are possible, and possibly useful.
The second component provided by the new statistics plugin, is the MatrixResource, a BioResource for documents (e.g. files on the harddisk) that represent a matrix. However, Bioclipse can create such matrices on the fly too, and these do not necessarily have to be stored on disk, as is general for BioResource’s. This makes it possible for other plugins to create matrices from other resources: for example, the CDK plugin can now have an action that converts a SDF file into a QSAR data matrix.
The MatrixResource can be edited using a plain text editor, and a more visually attractive graphical editor based on the KTable SWT widget:
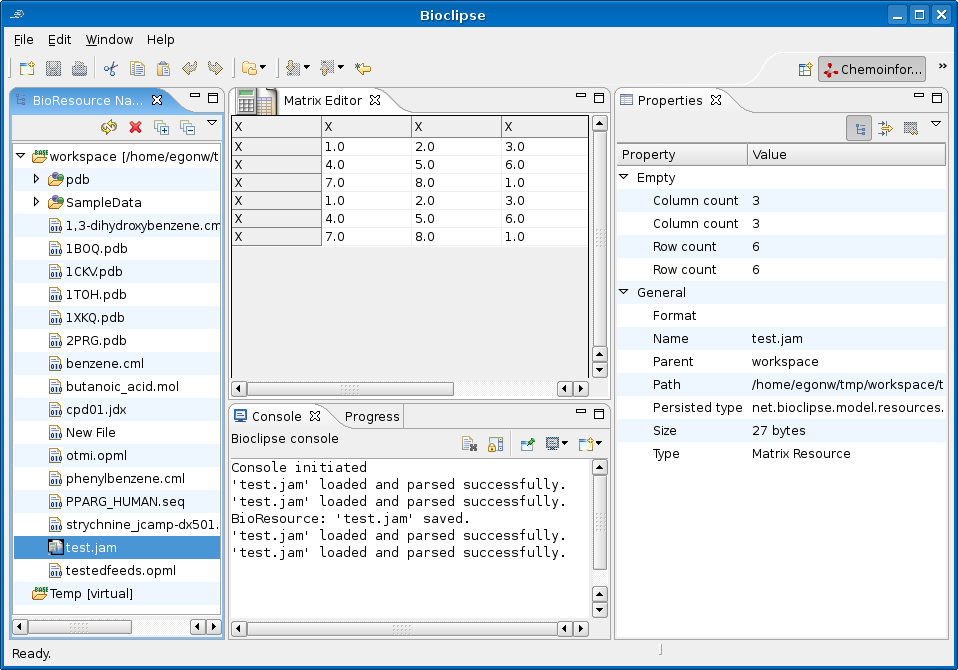
The next step is to work on column and row names, and replace those uninformative X’s. As you can see in the Properties View, I also need to tweak adding and removing advanced properties a bit. And then it is time to have the CDK plugin create a QSAR data matrix.