-
Converting JSON to RDF/XML with Groovy
Mark’s new CCO/RDF hosting functionality (see also my post two days ago) requires RDF/XML format, so I updated my code to convert the Chempedia Substances data into RDF/XML instead of N3 (I have asked Rich to put a new download link online). This is the Groovy code I used: -
Oscar: training data, models, etc
Oscar uses a Maximum Entropy Markov Model (MEMM) based on n-grams. Peter Corbett has written this up (doi:10.1186/1471-2105-9-S11-S4). So, it basically is statistics once more. If you really want a proper bioinformatics education, so do your PhD at a (proteo)chemometrics department. -
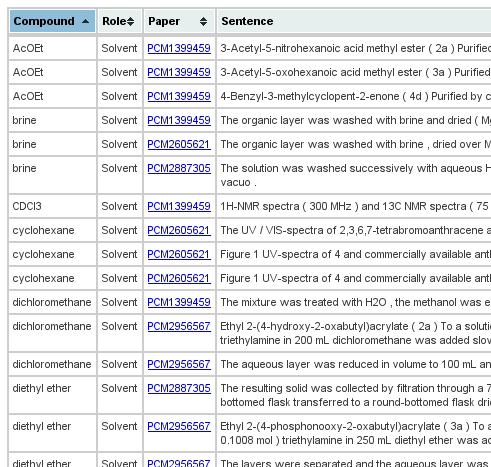
Status update on BJOC analysis with Oscar and ChemicalTagger #3
The two earlier posts in this series showed screenshots of results of Oscar, but the title also promised results by Lezan’s ChemicalTagger. Sam helped with getting the HTML pages online via the Cambridge Hudson installation. Where Oscar find named entities (chemical compounds, processes, etc), ChemicalTagger finds roles, like solvent, acid, base, catalyst. Roles are properties of chemical compounds in certain situations. Ethanol is not always a solvent, sometimes it is a Xmas present. The current output is not entirely where I want to go yet, but makes it easy which solvents are frequently found in the BJOC corpus: -
Adding a new dictionary to Oscar
Say, you have your own dictionary of chemical compounds. For example, like your company’s list of yet-unpublished internal research codes. Still, you want to index your local listserv to make it easier for your employees to search for particular chemistry you are working on and perhaps related to something done at other company sites. This is what Oscar is for. -
Oscar4 command line utilities
One goal of my three month project is to take Oscar4 to the community. We want to get it used more, and we need a larger development community. Oscar4 and the related technologies do a good, sometimes excellent, job, but have to be maintained, just like any other piece of code. To make using it easier, we are developing new APIs, as well as two user-oriented applications: a Taverna 2 plugin , and command line utilities. The Oscar4 Java API has slightly evolved in the last three weeks, removing some complexity. In this post, I will introduce the command line utilities. -
CiteULike CiTO Use Case #1: Wordles
Last month I reported a few things I missed in CiteULike. One of them was support for CiTO (see doi:10.1186/2041-1480-1-S1-S6), a great Citation Typing Ontology.