-
Programming in the Life Sciences #1: a six day course
Our department will soon start the course Programming in the Life Sciences for a group of some 10 students from the Maastricht Science Programme. This is the first time we give this course, and over the next weeks I will be blogging about this course. First, some information. These are the goals, to use programming to: -
CiteULike adds a HTML widget to embed citations
Spjuth, O.; Carlsson, L.; Alvarsson, J.; Georgiev, V.; Willighagen, E.; Eklund, M. Current Topics in Medicinal Chemistry 2012, 12, 1980-1986. -
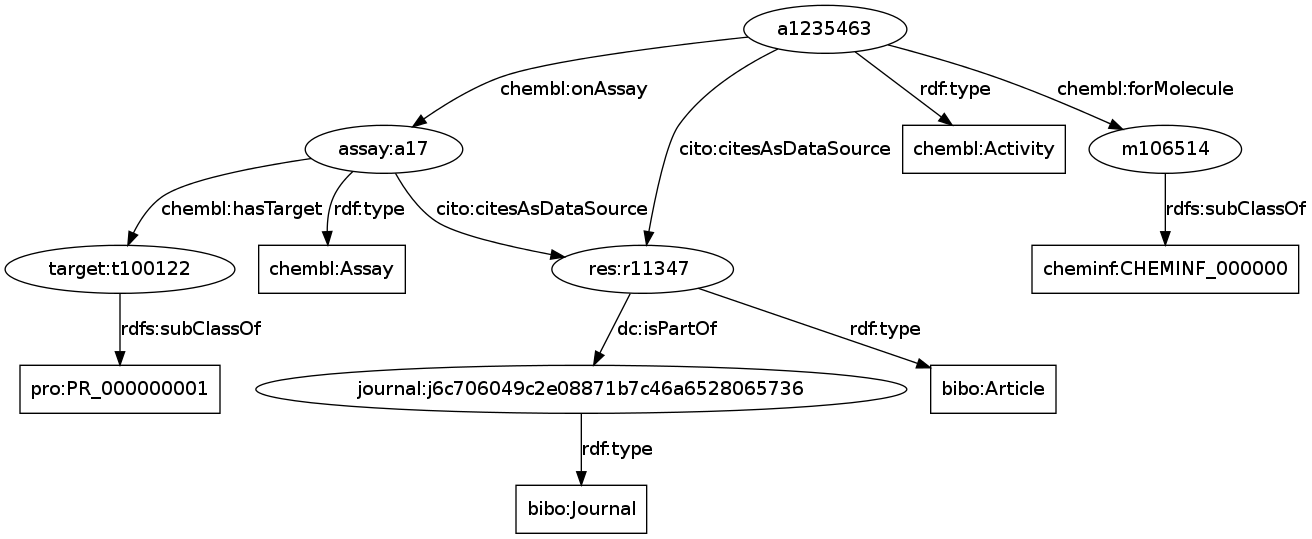
"Emerging practices for mapping and linking life sciences data using RDF"
The “Emerging practices for mapping and linking life sciences data using RDF” (doi:10.1016/j.websem.2012.02.003) is now available online, where I contributed a section on the original workflow for creating ChEMBL triples, and contributed to the section about open licensing, referring to CCZero and the Panton Principles. Happy reading! -
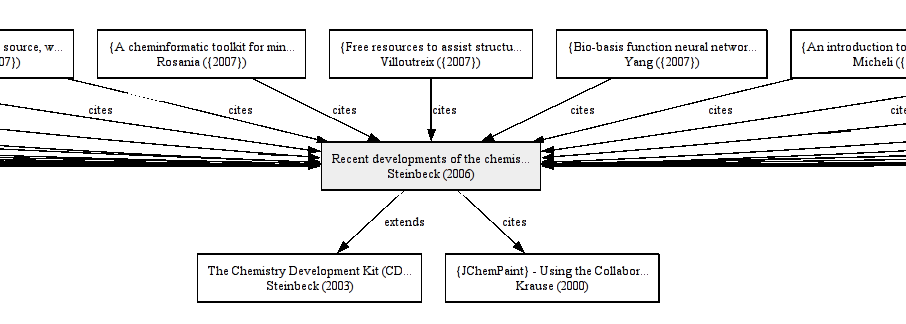
CiTO / CiteULike: publishing innovation
Readers of my blog know I have been using the Citation Typing Ontology, CiTO (doi:10.1186/2041-1480-1-S1-S6). I allows me to see how the CDK is cited and used . CiteULike is currently adding more CiTO more functionality, which they started doing almost one and a half years ago. -
Groovy Cheminformatics 4th edition
Six month was not quite the amount of time I anticipated between the third and fourth edition, but I finally managed to upload edition 1.4.7-0 of my Groovy Cheminformatics book. The first three editions sold 37 copies, including two for myself. Enough to feel supported and to continue working on it.