Where do the WikiPathways come from?
WikiPathways was founded in 2008, in the year I left Wageningen (and we Nijmegen) and moved to Uppsala, Sweden. When we dediced to move back to The Netherlands in 2012, I got to opporunity to join the Department of Bioinformatics (BiGCaT) and work on Open PHACTS. I had visited the group in March 2011 because I had a COST action workshop near Maastricht (about nanoQSAR) and the bioinformatics group did WikiPathways.
When I joined, there were already hundreds of pathways, originating from various collaborations (see below).
Around the winter break, the question came up who are the people who have drawn all these pathways. And on the new website
this is not actually that easy to see. You can browse all pathways, or look up
author profiles, but not all authors have done the same amount of work.
Moreover, at various points of time, batches of pathways from those collaborators were added. Often, these were added
by the MaintBot account, which is routinely hidden, and then the author who shows up as first author, is not even
the original author. And then we still have a lot of homology-converted pathways. These are pathways translated to
some species from a model species. You can find them in this repository.
But nowadays I do a lot in the WikiPathways project, among other things generate the RDF and maintain the code that does so. And I realized that we have author information in the RDF too (created by Alex Pico. So, the idea came up to see who the “first authors” are of the WikiPathways (mind the MaintBot issue), and what we know about them. Many already had their ORCID profiles linked from their profile pages, making it easy to look up their expertises.
Now, that was in January. But it turned out that the author information in the RDF worked fine in the .ttl file
of a single pathway, but that the series ordinal (e.g. 1 for being first author) was bound to the author, and
a SPARQL query would not be able to figure out on which pathways someone was first author. I fixed this somewhere
in January, so in the February 10 release the
improved data model was available.
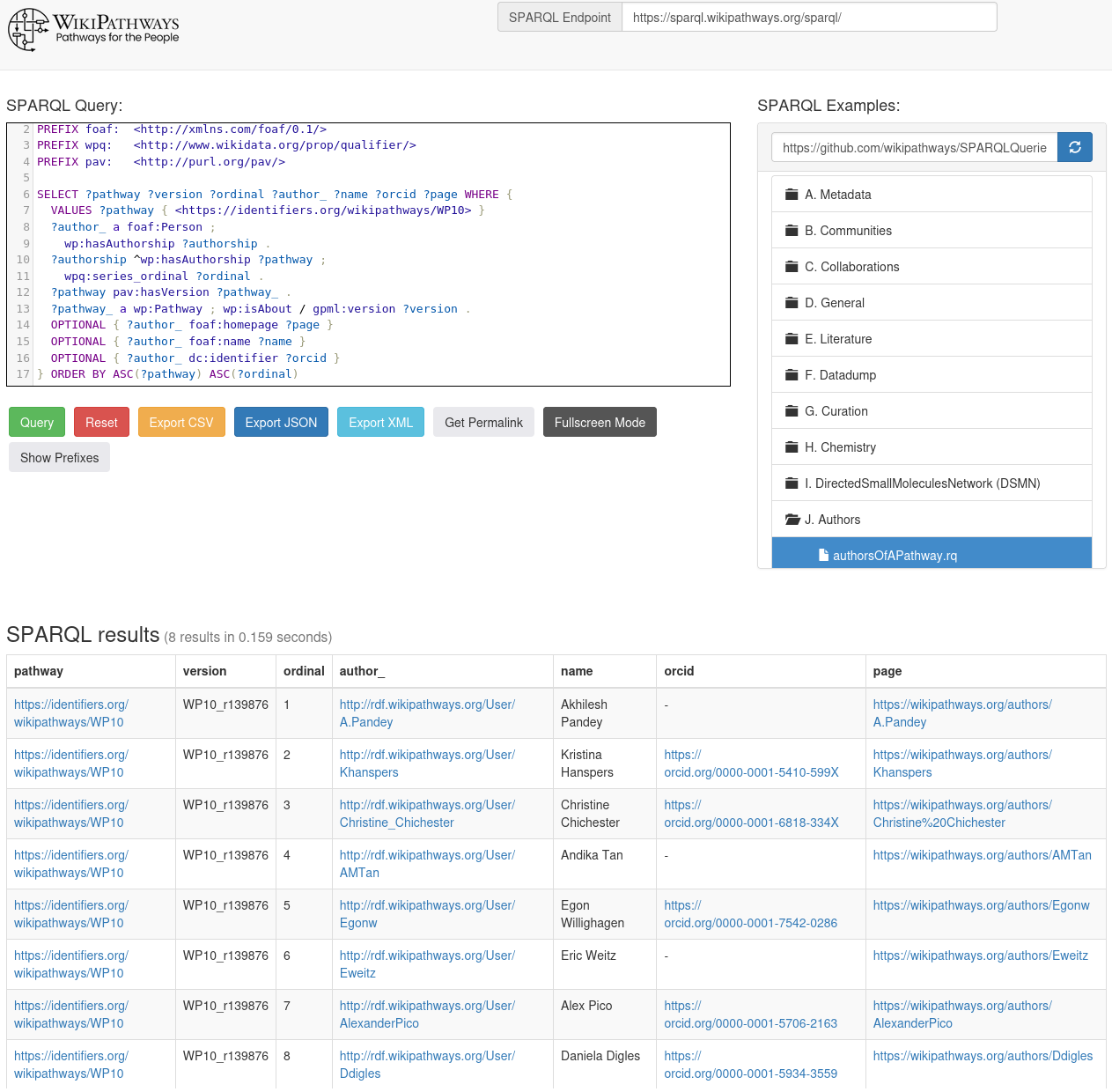
Allow me to show what is now possible, with a few SPARQL queries. First, list the authors of a pathway, use
this template for WP10:
PREFIX dc: <http://purl.org/dc/elements/1.1/>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
PREFIX wpq: <http://www.wikidata.org/prop/qualifier/>
PREFIX pav: <http://purl.org/pav/>
SELECT ?pathway ?version ?ordinal ?author_ ?name ?orcid ?page WHERE {
VALUES ?pathway { <https://identifiers.org/wikipathways/WP10> }
?author_ a foaf:Person ;
wp:hasAuthorship ?authorship .
?authorship ^wp:hasAuthorship ?pathway ;
wpq:series_ordinal ?ordinal .
?pathway pav:hasVersion ?pathway_ .
?pathway_ a wp:Pathway ; wp:isAbout / gpml:version ?version .
OPTIONAL { ?author_ foaf:homepage ?page }
OPTIONAL { ?author_ foaf:name ?name }
OPTIONAL { ?author_ dc:identifier ?orcid }
} ORDER BY ASC(?pathway) ASC(?ordinal)
We can see who the 8 people are who contributed to this pathway (we cannot actually see here what they contributed), and many authors are member of the WikiPathways review team who focus more on technical quality and the biology. The first author, however, often is the person who contributed most of the biological knowledge in the pathway, in this case Akhilesh Pandey from the NetSlim collaboration (see doi:10.1093/database/bar032):
Collaborations
Over time, multiple collaborations have taken place, like the one with NetSlim from the above query. In these collaborations, the knowledge may not be digitized in WikiPathways as GPML by the biological experts. That encoding regularly is done by others, but with those experts ensuring the quality. The following collaborations are examples, and a fuller list is found online:
- WormBase (doi:10.1093/nar/gkt1063)
- LIPID MAPS (doi:10.1093/nar/gkad896)
- Inherited Metabolic Disorders (doi:10.1007/978-3-030-67727-5_73)
- Micronutrients (doi:10.1007/s12263-010-0192-8)
We have collaborated with Reactome on various occassions (e.g. see doi:10.1371/journal.pcbi.1004941 and doi:10.1007/s12263-010-0192-8), around plants (e.g. see doi:10.1186/1939-8433-6-14), around rare diseases in projects like EJP-RD and ERDERA, and around SARS-CoV-2. For that, see these communities:
And then there are pathways in WikiPathways supported by a full paper, but I will leave that for a later moment.
Author statistics
Back to the authors, because the new RDF model allows a few more nice queries. For example, we can check the number of pathways with a certain number of authors, and then we find with the following query that there are two pathways with up to 18 authors (try here):
PREFIX dc: <http://purl.org/dc/elements/1.1/>
PREFIX wpq: <http://www.wikidata.org/prop/qualifier/>
SELECT ?atLeast (COUNT(DISTINCT ?pathway) AS ?count) WHERE {
?author_ a foaf:Person ;
wp:hasAuthorship ?authorship .
?authorship ^wp:hasAuthorship ?pathway ;
wpq:series_ordinal ?atLeast .
} GROUP BY ?atLeast
ORDER BY ASC(xsd:integer(?atLeast))
We can also look at the list of authors, sorted by the number of pathways they are noted as first author on. allong with their profile page on ORCID number:
PREFIX dc: <http://purl.org/dc/elements/1.1/>
PREFIX foaf: <http://xmlns.com/foaf/0.1/>
PREFIX wpq: <http://www.wikidata.org/prop/qualifier/>
PREFIX pav: <http://purl.org/pav/>
SELECT (COUNT(DISTINCT ?pathway) AS ?count) ?name ?orcid ?page WHERE {
VALUES ?ordinal { "1" }
?author_ a foaf:Person ;
wp:hasAuthorship ?authorship .
?authorship ^wp:hasAuthorship ?pathway ;
wpq:series_ordinal ?ordinal .
?pathway pav:hasVersion ?pathway_ .
?pathway_ a wp:Pathway ; dcterms:identifier ?version .
OPTIONAL { ?author_ foaf:homepage ?page }
OPTIONAL { ?author_ foaf:name ?name }
OPTIONAL { ?author_ dc:identifier ?orcid }
} GROUP BY ?ordinal ?name ?orcid ?page
ORDER BY DESC(?count)
Is this the full story? No, of course not. There are so much details yet uncovered, but it gives a bit more insight of where the biological knowledge in WikiPathways is coming from.
Want more peer review of the content? Then why not help setup a new community? Just ping me or Martina.
You can use your Mastodon or other ActivityPub-based Fediverse-account to comment on this article by leaving a publicly visible reply to this associated post. Content warnings are supported. You can also delete your comments at any time by deleting your post on the Fediverse.