PlantMetWiki: a linked open data service for querying and analyzing plant pathway knowledge
Back on October I presented Everything you always wanted to know: plant pathway modelling in WikiPathways (doi:10.5281/zenodo.18149988) at the Knowledge Graphs for Plant and Microbiome Multiomics symposium (see this archived LinkedIn post) on 14th October 2025 (youtube recording). I had not found time yet to post about this meeting, but it was an awesome list of speakers, regrettable absense of some others, but resulting in new contacts and some slowly evolving collaborations.
Previously, plant pathways were somewhat negatively prioritized at our BiGCaT research group. Something with Dutch academic politics. But that was 10 years ago, and with the notion that human health very much involves the exposome, which includes live around humans, I think the plant pathway science is important to human health. Even just the human health impacts of drops in biodiversity. Or the impact on our nutrition supply chain of climate change.
Anyway, I am happy that Elena and Denise pulled me into a collaboration to create an RDF-based knowledge graph about plant pathways. Their idea was to PlantCyc pathways (their license seems to allow that; doi:10.1093/nar/gkae991), convert that to GPML (by Max) and then to RDF. That last step is where I come in. The details will follow later, but Elena announced the project on LinkedIn (archived link), so time to blog about it myself too.
I am happy with this effort, not just because we now have pathways in RDF form for more than 500 species, but also because it requires continued development of the WikiPathways solutions, like GPML and libGPML and the RDF generation code, but also BridgeDb (doi:10.1186/1471-2105-11-5). The latter provides the identifier mapping infrastructure, but needed to be extended for the new species (something I had to do earlier this year for several caffeine synthesis pathways developed at the DBCLS BioHackathon 2025).
Lars gave me a tip on how to scale this up (after a manual addition), verifier.globalnames.org (doi:10.5281/zenodo.17245658, which greatly helped me out. It translates species names into identifiers, and their JSON is very rich in that process as well as easy to process. So, a custom script allowed me to update BridgeDb more efficiently. Highly recommended!
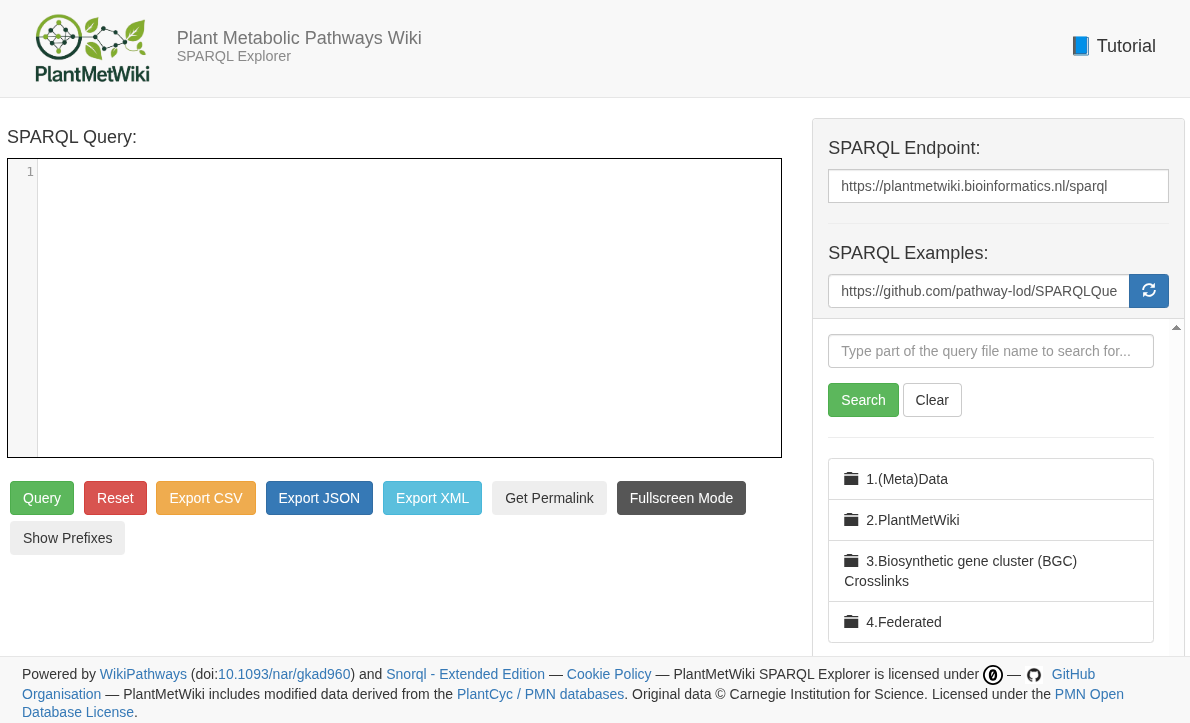
So, the resulting knowledge base is available at plantmetwiki.bioinformatics.nl and looks like this (also big thanks to Marvin for support in setting this up!):
You can use your Mastodon or other ActivityPub-based Fediverse-account to comment on this article by leaving a publicly visible reply to this associated post. Content warnings are supported. You can also delete your comments at any time by deleting your post on the Fediverse.