PFAS in the blood of the Dutch population
A recent report by the Dutch RIVM, PFAS in the blood of the Dutch population (doi:10.21945/RIVM-2025-0094), writes that seven PFAS compounds are found in blood samples of all tested people. Another nine compounds are found in at least 1-in-10 people. Because there is relevant data in the report on the 28 studied PFAS compound, I wanted to have the report more FAIR than it is on the website. Why this report? Well, the chemistry and the history is fascinating and brutal (I like this Veritasium video).
The history tells me that our society may sound woke and leftish, in reality it is a continous fight for basic human rights. (Something that plenty have been saying for years.) In this case, a healtht life is the human right.
So, what can I do to make this report more FAIR?
Findable in Wikidata
Since this report has been mentioned in the news, it clearly is notable. The simplest thing to do is thus to just add it Wikidata. Because the DOI of the report had not been recorded yet, I could not let Scholia do it for me. But doing it manually is only a bit more work: Q135222054. The provided metadata on the RIVM website is minimal.
But we can do more. Particularly, because I want people to find this report when they look info knowledge about the 28 studied chemicals, I added main subject annotation using the information in Table 1 in the report. Using Scholia and the CAS registry number in the table, I crosscheck the information in Wikidata is consistent with the report (and visa versa). It was. I then added the Dutch name and acronym for most of them. Some already had the name as in the Table. That gives us a nice “Topic scores” plot for the Scholia page of the report:
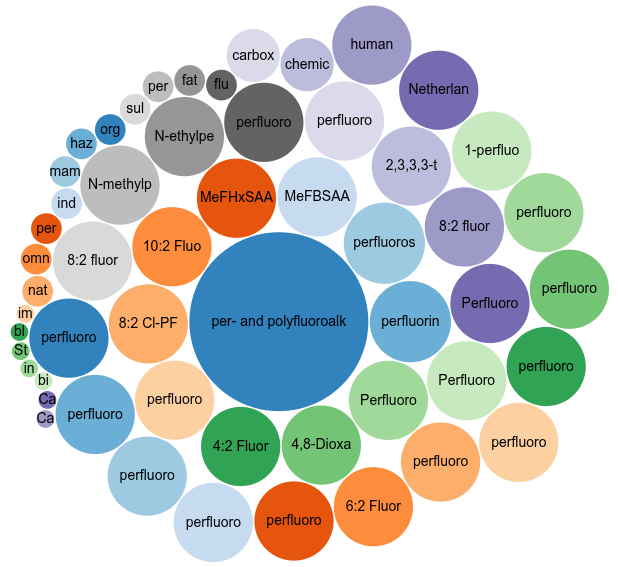
The central PFAS bubble is also only one main subject but larger because many the specific PFAS compounds are subclassing PFAS. And you may also note many smaller bubbles. These actually come from main subject annotations of articles cited from the report. Because I added a few of them too. Not all, because many are not in Wikidata (yet).
Findable in WikiPathways
But since 16 of these compounds are readily found in human blood samples, that is handy knowledge when doing metabolomics (on blood samples). Or (and I leave that to later blog post), we can map the experimental data for Dordrecht versus the rest of The Netherlands to the PFAS compounds. That is relevant to research by VHP4Safety. There are many ways to see if you have PFAS in your dataset, but since we have many controlled lists of genes in metabolites, I added one for common PFAS in human blood samples. Well, the 16 common in Dutch blood samples:
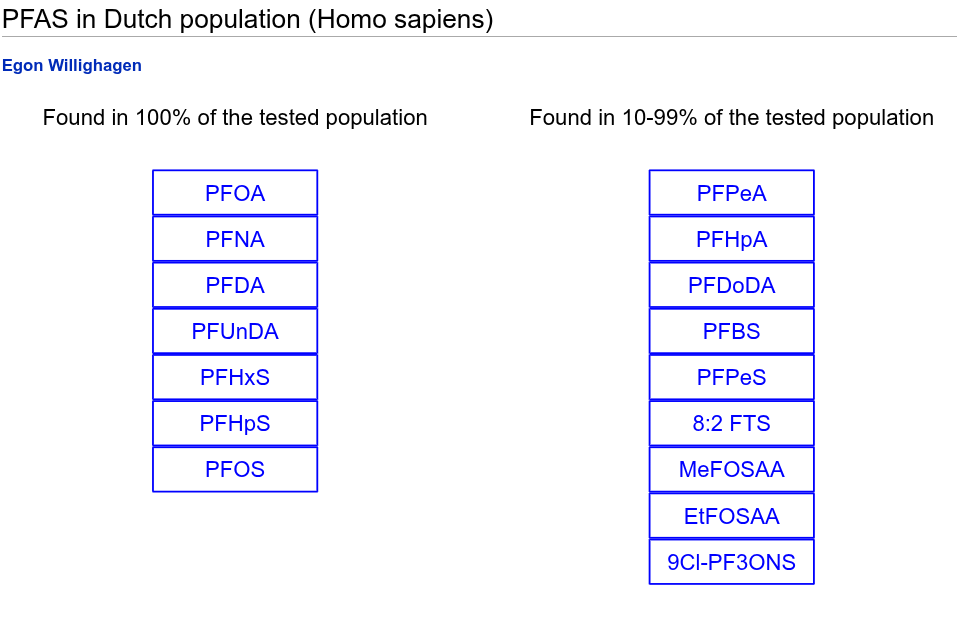
Each metabolite here is annotated with their Wikidata identifier, allowing us to map experimental data on top of it. And we get links out to other databases almost for free:
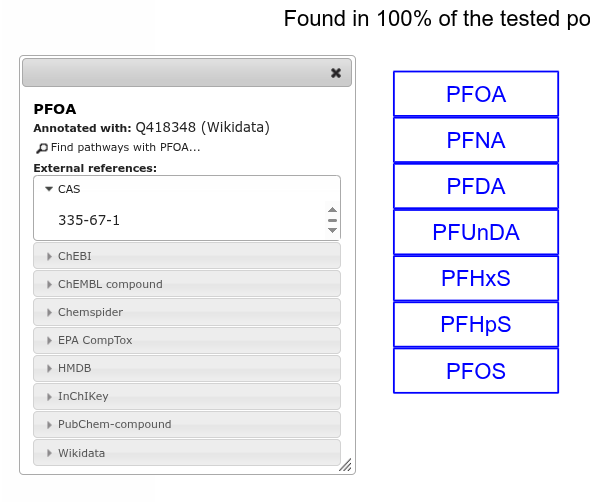
And the link to Wikidata actually links to Scholia, so for the PFOA in the above example, we can quickly see the boiling point, decomposition point, and melting point of this PFAS. And literature with undoubtedly even more knowledge about this PFAS:
Now, these two steps were mostly manual: drawing WP5579 in WikiPathways and adding the report annotations (main subject and cites) in Wikidata.
Findable in the VHP4Safety Compound Wiki
As part of the VHP4Safety project, I am collecting information on chemicals studied in the context of toxicology, safety, and risk assessment. Often specific collections of compounds studied as a whole. This report is such a collection and provides experimental data on these compounds. So, I want this report to be findable for the VHP4Safety Compound Wiki too. Creating the collection is a manual step: Q5145.
Now, because both Wikidata and our VHP4Safety Compound Wiki (a Wikibase instance) are semantic and support, I can use SPARQL to create instructions to link the 28 compounds to the new collection. Now, arguably, that can be done manually too, and maybe faster, for larger collections this is harder. So, I dug up my earlier notes and got some useful things together.
This query lists all 28 PFAS linked to the report in Wikidata:
SELECT ?pfas ?pfasLabel WHERE {
wd:Q135222054 wdt:P921 ?pfas .
?pfas wdt:P31 wd:Q113145171 .
SERVICE wikibase:label { bd:serviceParam wikibase:language "[AUTO_LANGUAGE],mul,en". }
}
Using federation powers, I can use this for a SPARQL query, and return the results in QuickStatements that say this VHP compound is part of the VHP collection:
PREFIX wb: <https://compoundcloud.wikibase.cloud/entity/>
PREFIX wbt: <https://compoundcloud.wikibase.cloud/prop/direct/>
SELECT (SUBSTR(STR(?cmp),45) AS ?qid) ?P21 WHERE {
?cmp wbt:P5 ?wikidata .
SERVICE <https://query.wikidata.org/sparql> {
wd:Q135222054 wdt:P921 ?pfas .
?pfas wdt:P31 wd:Q113145171 .
BIND (substr(str(?pfas),32) AS ?wikidata)
}
BIND ("Q5145" AS ?P21)
}
I actually had to add 5 PFAS compounds in the VHP4Safety Compound Wiki first. That follows the
same procedure for how I have been adding chemical compounds to Wikidata
(see also this preprint).
The input cas.smi has the (missing) SMILES, Wikidata QID, and English label:
C(CS(=O)(=O)O)C(C(C(C(C(C(F)(F)F)(F)F)(F)F)(F)F)(F)F)(F)F Q27063662 6:2 Fluorotelomer sulfonate
CN(CC(=O)O)S(=O)(=O)C(C(C(C(C(C(F)(F)F)(F)F)(F)F)(F)F)(F)F)(F)F Q126605979 MeFHxSAA
CN(CC(=O)O)S(=O)(=O)C(C(C(C(F)(F)F)(F)F)(F)F)(F)F Q126682412 MeFBSAA
C(=O)(C(C(F)(F)F)(F)OC(C(C(F)(F)F)(F)F)(F)F)O[H] Q29387971 2,3,3,3-tetrafluoro-2-(heptafluoropropoxy)propanoic acid
C(C(C(=O)O)(F)F)(OC(C(C(OC(F)(F)F)(F)F)(F)F)(F)F)F Q81981675 4,8-Dioxa-3H-perfluorononanoic acid
For reference, this is the command line I used to create QuickStatement instructions:
groovy createWDitemsFromSMILES.groovy -w compoundcloud.wikibase.cloud -c Q2368 -d P5 -l -i wikidata -a P11
Final remark
Are these 16 the only PFAS in our body? With 28 studied out of a potential seven million, I doubt it.
You can use your Mastodon or other ActivityPub-based Fediverse-account to comment on this article by leaving a publicly visible reply to this associated post. Content warnings are supported. You can also delete your comments at any time by deleting your post on the Fediverse.