Niche papers and citation intentions
I wish I could say I remember the first citation to one of my research articles. I do not. But I do remember the excitement to see why someone was citing my research. What I do remember is that I got a comment around the same time along the lines of this: “why would anyone cite your article if they can download the results for free?” (about open science cheminformatics research). Other times. Indeed, I found out there are many reasons why people are citing and not citing articles. The above is one of them (still happens too often). But that’s also an intrinsic property of the current publishing model: some papers get cited too much, others get cited too little.
Mark Dingemanse wrote up a post [i]n praise of niche papers, suggesting people to highlight papers that are not cited enough (as proxy for not getting enough attention). They write:
Let’s define niche papers informally as work to be proud of even if it managed to remain a bit obscure; good work that would deserve more readers. Niche papers may not contain the most flashy results. They may not appear in the most glamourous venues. They may be book chapters. They don’t easily gather drive-by citations.
Why I found this post interesting
Before I move on to highlighting niche papers (from our group and from others), I want to ponder a bit more about the rest. The first I learned is that the citation count to articles is a bad measure for the impact (2006 pondering): articles using your work may get more citations than your own article. For example, the first paper (doi:10.1021/CI025584Y) about the open science cheminformatics about the Chemistry Development Kit (CDK) was originally cited less than the paper about the BRENDA enzyme database (doi:10.1093/NAR/GKH081) using the CDK for fingerprint calculations (to compare and search enzyme substrates), and later much less than MZmine (doi:10.1186/1471-2105-11-395) (see this Scholia page):
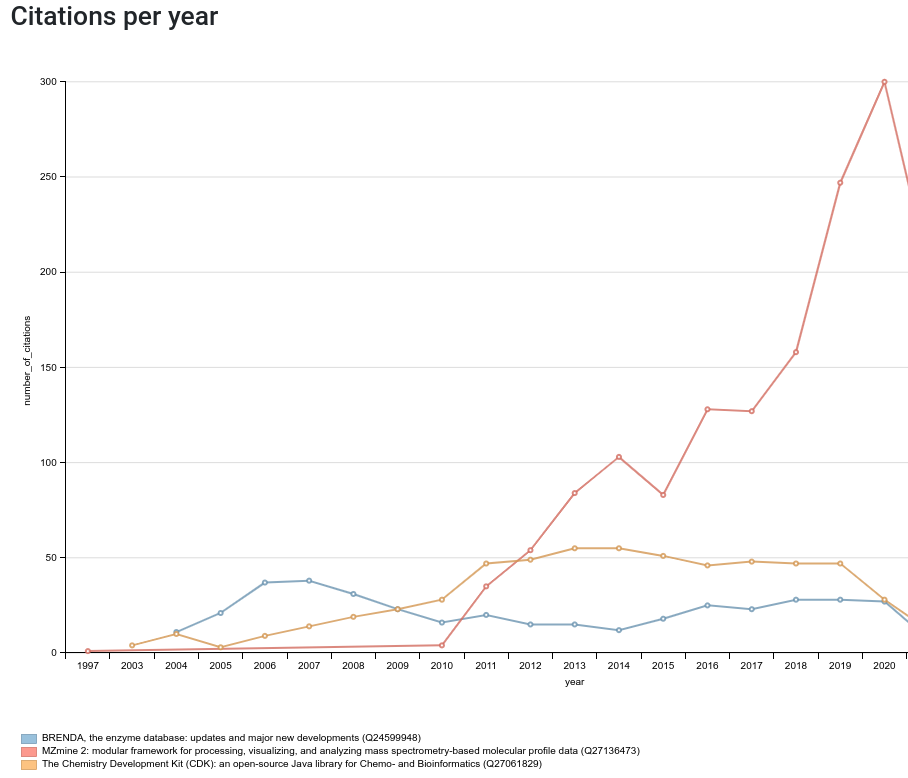
I think we should with limiting ourselves to papers and book chapters. We must extend out notion of research output, anyway, starting with data and software. This is part of defining what niche is, imo.
Second reason why I liked Mark’s post is the drive-by citations, which he references to a 2009 post by andrewperrin which defined such a citations as
references to a work that make a very quick appearance, extract a very small, specific point from the work, and move on without really considering the existence or depth of connection between the student’s work and the cited work.
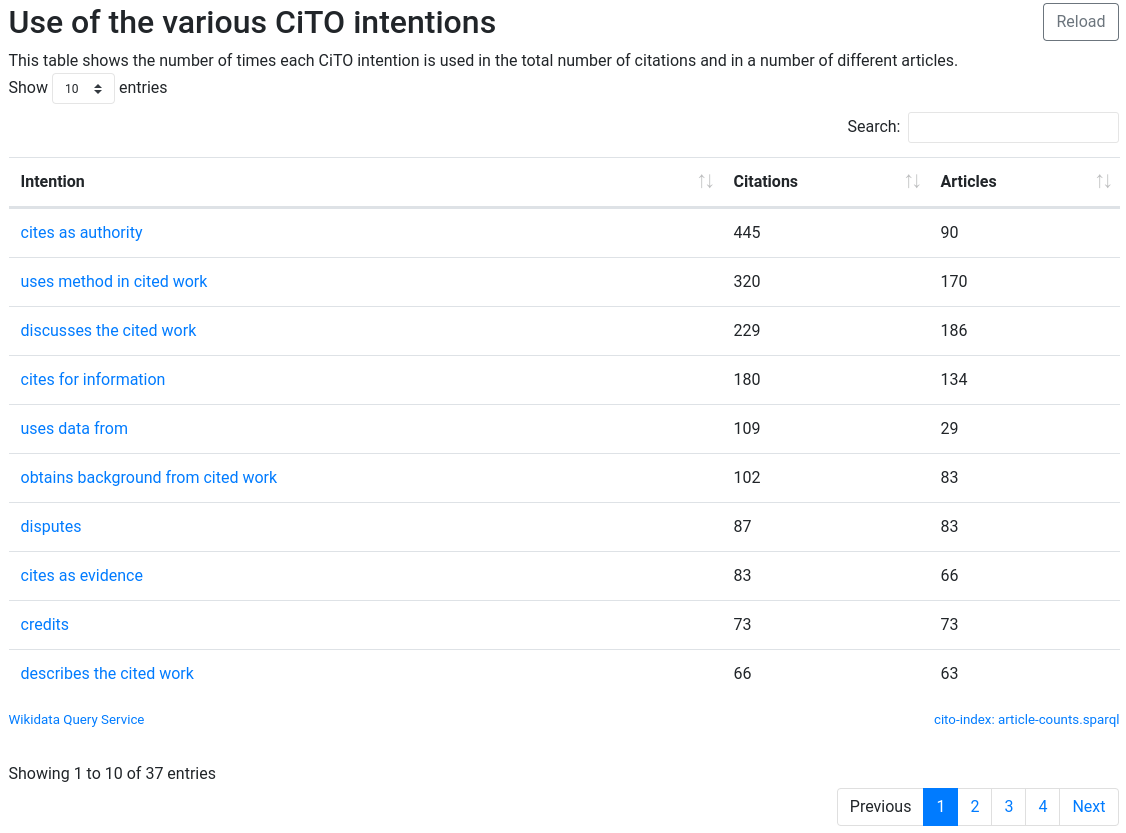
This is someone I noted too when analyzing citations to the aforementioned CDK paper. Particularly in the early days, it was cited a lot in a similar way: it was not using the CDK, but ascribed some authority to the paper in a very quick appearance, without really considering the cited work. The Citation Typing Ontology (CiTO, doi:10.1186/2041-1480-1-s1-s6) has cito:citesAsAuthority for that (not exactly the same thing, and maybe CiTO should have cito:driveByCitation too). And they happen a lot, and in the past I have guestimated them to make up 20-35% of the citations to an article, and I postulate that high-journal-impact-factor journals amass a higher ratio than specialistic (niche?) journals.
With FAIR citations (see this post) we can visualize that ratio, here in this Scholia page:
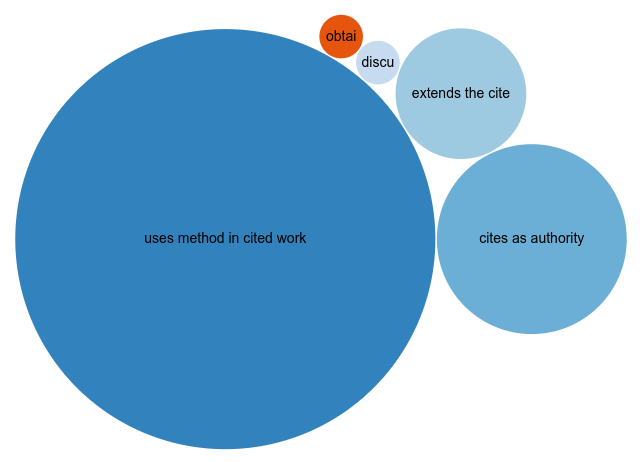
It is also obvious that the first CDK paper introduced a new method. But the pattern is not limited to this paper, and with just over 2000 citation intentions, we start of get some idea of this pattern:
My contributed Niche Papers
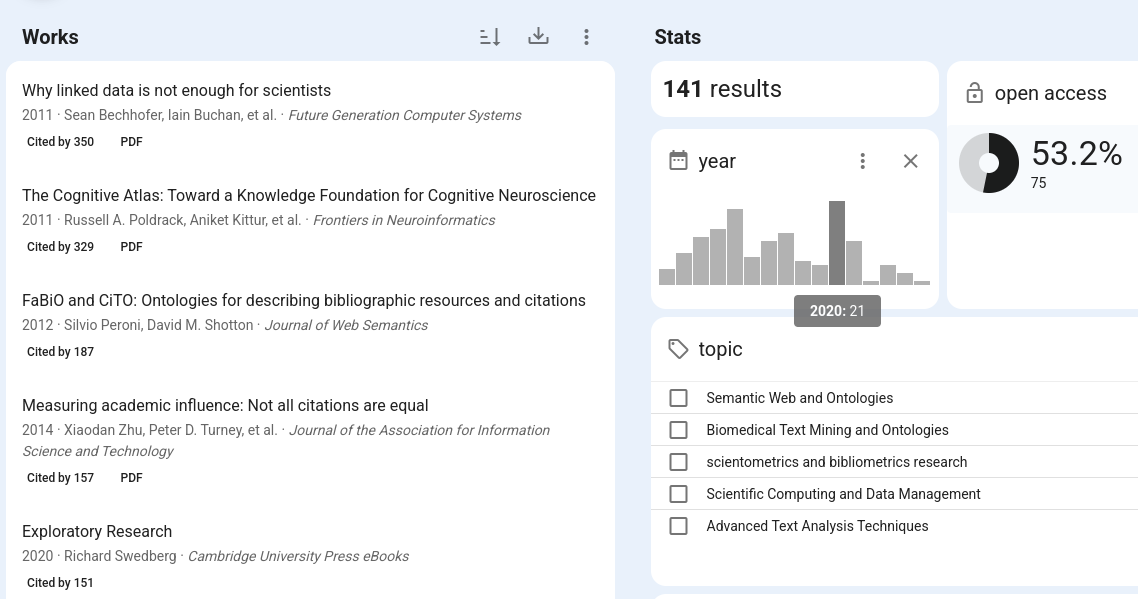
That brings me to a first neglected paper, David Shotton’s original conference proceedings CiTO, the Citation Typing Ontology (doi:10.1186/2041-1480-1-S1-S6), another paper where citing articles are more cited than the original:
A second example is cited even less (only 36 times according to OpenAlex), but a wonderful early example of machine learning of a massive amount of data: Genome‐Scale Classification of Metabolic Reactions: A Chemoinformatics Approach (doi:10.1002/anie.200503833) by Diogo Latino and João Aires‐de‐Sousa. My 2006 blog post about their article did not make a difference. And this is remarkable if you look at home many articles are published now yearly in similar efforts.
From our group, I think the impact of Ryan Miller’s Understanding signaling and metabolic paths using semantified and harmonized information about biological interactions (doi:10.1371/journal.pone.0263057 is not fully appreciated yet. This paper describes and validates work by Ryan, Martina Kutmon, Answesha Bohler, and Andra Waagmeester on modelling biological interaction in a FAIR way. It builds on earlier work, like the WikiPathways RDF work by Andra (doi:10.1371/journal.pcbi.1004989), but zooms in on the interactions and develops method to assess the quality of the FAIR modelling of them. This provides us with a method to evaluate later analyses where these interactions are used.
A second paper from our group which I expected to get more attention is a paper by Ammar (doi:10.1186/s13321-023-00701-3) where he looked into personalized binding affinities. That is, drugs may bind better to their targets for some people than for other (and therefore work better for some people than for other), and his analysis suggests they impact can be significant. We will learn in time.