Serious Request: the results
The last week before the winter break Serious Request took place. We started an action around WikiPathways and we collected 877 euro for the MetaKids Foundation. In total there were 2612 actions, many of which brought in a lot more. We ended up in position 928.
But the money was only one part of our “donation” of the MetaKids goal to make 35 percent point more inherited metabolic disorders treatable (which they currently are not), and to address the number one cause of death among Dutch kids. Because our action focussed on getting more biology relevant to metabolic diseases into WikiPathays. For this we set up a WikiPathways SR24 community page, along with a curation page showing the results of automated curation alerts. Actually, in preparation of the Action, I updated that code base to no longer have two states (succeeded, failed), but four states, depending on the percentage of tests failing for that pathway. This has also been roled out to the main WikiPathways website.
In the weekend before our action, I wanted to test my PathVisio and had a go at a pathway drawing from a book of which most pathways had already been digitized (see doi:10.1007/978-3-030-67727-5_73), but not this one. This resulted in a first pathway (wikipathways:WP5504), which was later that week greatly extended by Denise. I also ported the table of chapters from this book to the new WikiPathways community page for the book.
A list of genes
From Marek Noga from our university medical center I received a pointer to a nice paper with a long list of diseases and matching genes (doi:10.1002/jimd.12348) which provided a great starting point. I started out by making the data from the supplementary files more FAIR by converting the data into RDF.
With SPARQL I compared the genes (via their HGNC symbols) with the content of WikiPathways:
PREFIX wp: <http://vocabularies.wikipathways.org/wp#>
PREFIX dc: <http://purl.org/dc/elements/1.1/>
SELECT ?gene ?omim ?geneLabel WHERE {
?gene a wp:GeneProduct ;
rdfs:label ?geneLabel .
OPTIONAL {
?gene rdfs:seeAlso ?omimIRI .
?omimIRI dc:identifier ?omim .
FILTER (contains(str(?omimIRI), "omim:"))
}
?gene wp:bdbHgncSymbol ?hgnc .
OPTIONAL {
SERVICE <https://sparql.wikipathways.org/sparql> {
?wpGene wp:bdbHgncSymbol ?hgnc .
}
}
FILTER (!BOUND(?wpGene))
FILTER (CONTAINS(?geneLabel, " "))
}
This resulted in a spreadsheet with more than 300 genes not in WikiPathways. An analysis by Karen Rothfels and Lisa Matthews showed that the number of genes not found in Reactome is only 129. Indeed, later analyses showed that Reactome has a few very relevant pathways missing in WikiPathways.
New biological pathways
To figure out, it turns out the Pathway Figure OCR (doi:10.1186/s13059-020-02181-2) and NDEX (doi:10.1093/bioinformatics/btad118) tools are very useful here. They both allow passing a list of genes and return results (sets, pathways, models) relevant to that list. NDEX includes the sets from Pathway Figure OCR, and those sets are a set of genes linked to single journal article which included a pathway diagram. I used this on the list of 371 genes not in WikiPathways and the list of 129 genes not in Reactome, and identified five articles. It actually turns out that two basically described the same biology and both are captured in the same new pathway (wikipathways:WP5505). This pathway includes a good number of PIG genes, handling the very specific metabolic conversion of a metabolite.
Complex chemistry
That metabolite is complex and databases do not seem to have the structure yet, so I set out generating a SMILES:
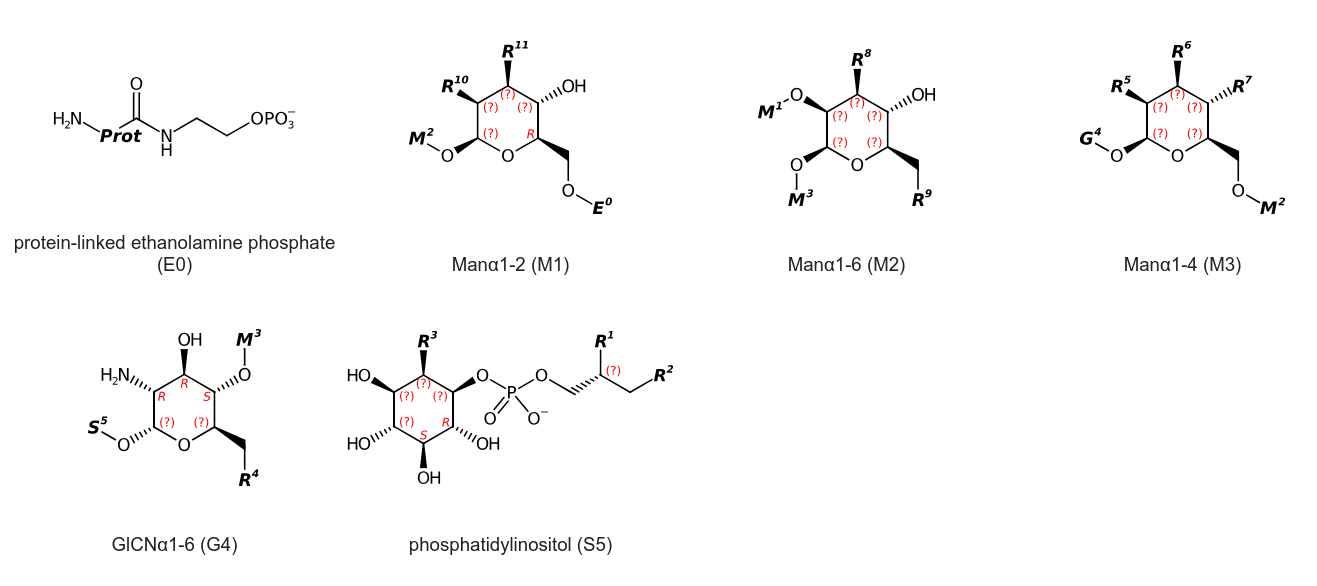
I reported the final SMILES, but I am not happy with it yet, and actually spotted an error already:
N[Prot]C(=O)NCCOP(=O)([O-])OC[C@@H]1[C@@H](O)[C@H]([R11])[C@H]([R10])[C@@H](O1)O[C@H]1[C@@H]([R8])[C@H](O)[C@@H](C[R9])O[C@H]1OC[C@@H]1[C@@H]([R7])[C@H]([R6])[C@H]([R5])[C@@H](O1)OC[C@@H]1[C@@H](O[M3])[C@H](O)[C@@H](N)[C@H](O1)O[C@@H]1[C@@H](O)[C@H](O)[C@@H](O)[C@@H]([R3])[C@H]1OP(=O)([O-])OC[C@H]([R1])C[R2]
So, for completeness and as backup, here are the fragment SMILES that you can copy/paste into CDK Depict:
N[Prot]C(=O)NCCOP(=O)([O-])OC[C@@H]1[C@@H](O)[C@H]([R11])[C@H]([R10])[C@@H](O1)O[C@H]1[C@@H]([R8])[C@H](O)[C@@H](C[R9])O[C@H]1OC[C@@H]1[C@@H]([R7])[C@H]([R6])[C@H]([R5])[C@@H](O1)OC[C@@H]1[C@@H](O[M3])[C@H](O)[C@@H](N)[C@H](O1)O[C@@H]1[C@@H](O)[C@H](O)[C@@H](O)[C@@H]([R3])[C@H]1OP(=O)([O-])OC[C@H]([R1])C[R2]
N[Prot]C(=O)NCCOP(=O)([O-])O protein-linked ethanolamine phosphate (E0)
[E0]OC[C@@H]1[C@@H](O)[C@H]([R11])[C@H]([R10])[C@@H](O1)O[M2] Manα1-2 (M1)
[M1]O[C@H]1[C@@H]([R8])[C@H](O)[C@@H](C[R9])O[C@H]1O[M3] Manα1-6 (M2)
[M2]OC[C@@H]1[C@@H]([R7])[C@H]([R6])[C@H]([R5])[C@@H](O1)O[G4] Manα1-4 (M3)
[R4]C[C@@H]1[C@@H](O[M3])[C@H](O)[C@@H](N)[C@H](O1)O[S5] GlCNα1-6 (G4)
[G4]O[C@@H]1[C@@H](O)[C@H](O)[C@@H](O)[C@@H]([R3])[C@H]1OP(=O)([O-])OC[C@H]([R1])C[R2] phosphatidylinositol (S5)
The hackathon day
On Thursday we had a hackathon day at our Translational Genomics department (UNS60 building). One of the Action organizers was still travelling back from Germany, but otherwise Tina, Denise, Daan, me, and Marek worked on Thursday on various things. Tina worked on WP5505, Daan created his first pathways (wikipathways:WP5507), and so did Marek (wikipathways:WP5506).
We now have 36 pathways on the community page:

After that hackathon, and to wrap up things, I finalized the updated to the curation page, making the output look better (more curation tests now output Markdown) and failing tests now almost all have an explanation page showing how the affected pathway can be improved (to address the issue).
Somewhere next week, the results of the pathways will be available from the WikiPathways SPARQL endpoint and I can then calculate new numbers. The number of genes not in WikiPathways should be lower.
Finally, perhaps, there are some very specific results, but also we have created a nice todo list:
- plenty of curation on those 36 pathways remains to be done
- we still have many genes of interest not in pathways, and we should start stubs in WikiPathways
- we need a better overview of the mitochondiral biology
And there are also still a few issues open:
- I have a todo item to make a curation SPARQL query available via the automated testing (enhancement)
- not all interactions end up in the RDF (bug)
That bug actually has significant impact on downstream analyses, I guestimate.
You can use your Mastodon or other ActivityPub-based Fediverse-account to comment on this article by leaving a publicly visible reply to this associated post. Content warnings are supported. You can also delete your comments at any time by deleting your post on the Fediverse.