Additional files, data, datasets, databases, and published data
Open Science doesn’t make publishing easier. That that’s all for the better: our research efforts are complex, so why should the publishing be. Sure, I am not talking about references formatting or moving the Methods section to the right location, or some silly statement that all authors agree with the manuscript when you are the only author.
No, let’s talk about data. What should you publish? How, and when? And why would you do it in the first place? This is not going to be a post about FAIR either, but instead about when to publish data as additional files (aka supplementary data), raw data, processed data, as a datasets, or even as a database. That’s a lot of types of data, and the differences matter at least for the effort you want to put in.
First, things have changed. We produce a massive amount more data. In the past your data, or at least the processed data, would be part of your conference talk, your journal article, or your book (chapter). Open Science has changed this: data should be easier to reuse. But that results in new questions; those as in the previous paragraph. So, let’s add some context.
Data is very broad and includes digital knowledge. Data can be raw, and the exact numbers collected (e.g. by a apparatus) or created by researchers. Processed data is what you get when you process the raw data. For example, raw data may be a FID graph in nucleic magnetic resonance, while processed data would be a plot showing intensities versus chemical shifts. Published data is then a list of peaks you put in your results section to support your claim of chemical identity.
A fourth type of data is metadata, and could here be the instrument on which the FID was measured, or the solvent used, etc. This is where it gets complicated, because depending on the researcher who processes the data, metadata can actually be data itself. For example, when you study the chemical shift differences in different organic solvents.
From a more social level, the Open Science 101 uses the following categories: primary data as collected/recorded by the researcher, and “secondary data typically refers to data that is used by someone different from who collected or generated the data”. This angle of data captures the collaboration aspects of open science, but says more about the processors than the data, I think.
Monitoring Open Data
Central aspect of doing research is to disseminate the research. Traditionally, this has been disseminating results, hoping they become facts. Increasingly, we realize that this process needs improvement, particularly clearly studies, done, and communicated by the Open Science approaches.
Complementary, there is recognition&rewarding (R&R) and the wish to use various kinds of monitoring to assess who should be rewarded (and who should be fired), and the monitor is the implementation of the recognition. So, how does this work for open data? We can count every open data, but if thrown on a big pile, that becomes a bad monitor for use in recognition and rewarding.
One idea is to differentiate in what data we monitor? Just raw data? Or processed data? How much intellectual effort does that have to in collecting/recording the data? Should that be part of the monitor and how do you even measure that? Lot’s of known unknowns here.
But this should not inhibit us from telling the research narative. And maybe we should just exploring the possible narratives to allow us how it may help us monitor work done, how to recognize contributions to the scientific record, and how to use all that in R&R.
I here present some example from my own research, just to start a narrative.
Raw data
Over the years I have collected and recorded quite a bit of raw data. First data collected in the lab and later mostly recorded. Even though I have been doing Open Science since the late nineties, I cannot say all my data has been archived well. Even less so, I do not have a “publication list” of all my raw data. As an academic community, we have been focusing too much on the scholarly article as the center of the research system (more on that later, because there is awesome research presented at the Dutch National Open Science Festival).
- pKa values (not archived, no DOI)
- NanoWiki 5 (archived, with DOI)
Processed data
As is defined in the European laws around GDPR, processing “includes the collection, recording, organisation, structuring, storage, adaptation or alteration, retrieval, consultation, use, disclosure by transmission, dissemination or otherwise making available, alignment or combination, restriction, erasure or destruction of [..] data”. As you can see, this is slightly different from the first, but in light of protecting citizen, this broader definition makes sense. My point here the that processing should be taken broadly. And data curation, which researchers routinely do, is processing too. For any data scientist, this is easily taking up 25% of the full time needed for any data analysis. One of the points of the FAIR principles is to keep that number as low as possible, but not really the point here.
When it comes to this kind of data, I like people to have readily access to the results of my curation. You will find a lot of processed data like this archived. Some examples of data by me or to which I contributed:
- WikiPathways (monthly archived, with DOIs)
- ChemPedia RDF (different format than original data, archived, with DOI)
- BridgeDb Metabolite ID mapping database (irregular releases, not every one is notable; archived, with DOI)
The last one will look something like this:
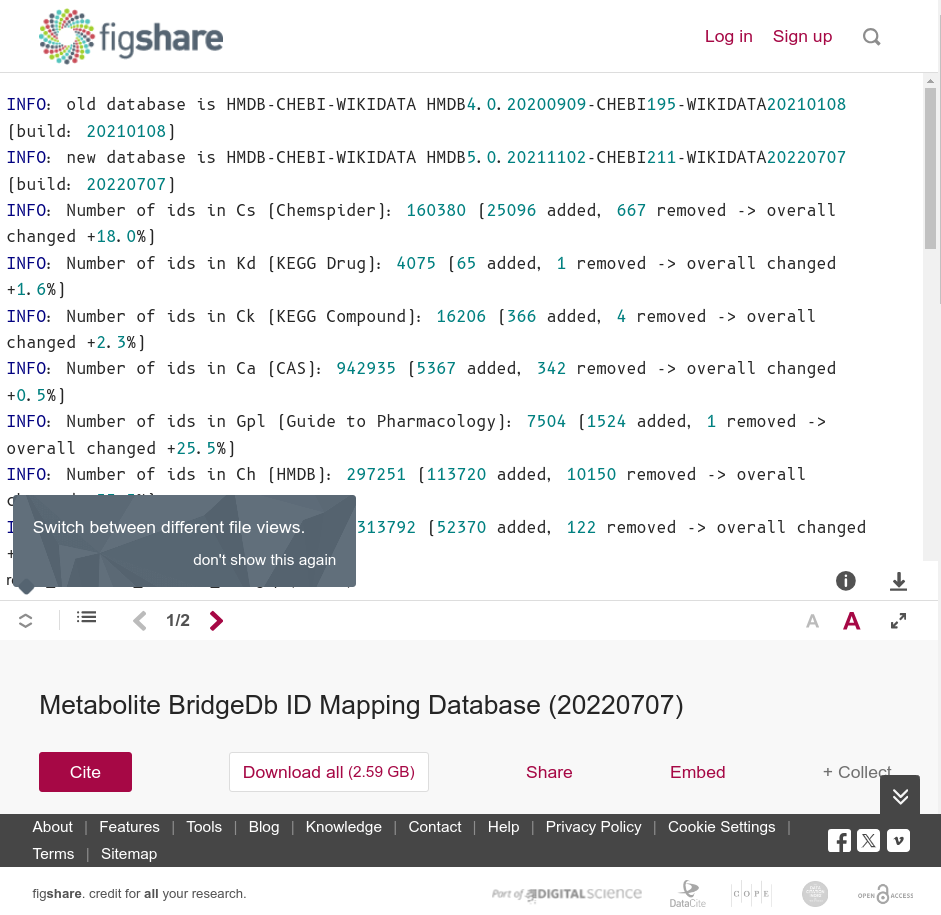
Published data
And then we have published data, which refers to data presented in a publication, like a journal articles. We know this as supplementary data or additional files. Several publishers, like BioMedCentral, submit these data automatically to a repository. For example, the Journal of Cheminformatics publishes all additional files under a CCZero license on Figshare. But many of these support the narrative of the story, rather than the narrative of the research question. Of course, journals also have limited expectations of the format and my personal impression is that these are not commonly FAIR. (Open Access is not Open Science.)
Some examples of such datasets where I do not see them as notable and do not expect them to be monitored. These datasets are part of the journal article, and that narrative is already monitored.
- MOESM1 of PubChemRDF: towards the semantic annotation of PubChem compound and substance databases (Word document with data, with DOI)
- MOESM1 of XMetDB: an open access database for xenobiotic metabolism (archived Structured Data file with chemical structures, with DOI)
Databases
And then we have databases provides as interactive website. This allows other researchers to explore the data, before the start processing the data. These typically do not have a DOI itself, tho data can be routinely archived as in the above WikiPathways example.
Databases itself, as research output, are much harder to archive. And to make them citatable, research publish journal articles with a narrative that describes the database. The follwing two are such database papers, where the article DOI is a proxy for the database:
- The ChEMBL database as linked open data (online, DOI via article)
- PSnpBind (online, DOI via article)