Scholarly discussions through the eyes of CiTO (and Wikidata)
Diabetes was already discussed in literature back in 1838-1839 (doi:10.1016/S0140-6736(02)96038-1, doi:10.1016/S0140-6736(02)96066-6, and doi:10.1016/S0140-6736(02)83966-6). These three papers show a short discussion. Papers were a lot shorter back in the days, and the discussion actually shows why papers are longer now (tho I am not sure they really got sufficiently more reproducible, but that’s another discussion).
Traditional citation counts do not make this discussion obvious, but if we make our publishing sufficiently FAIR (it’s far from that, right now), then we can get a step closer. For example, with the Citation Typing Ontology we can show how the papers relate to each other:
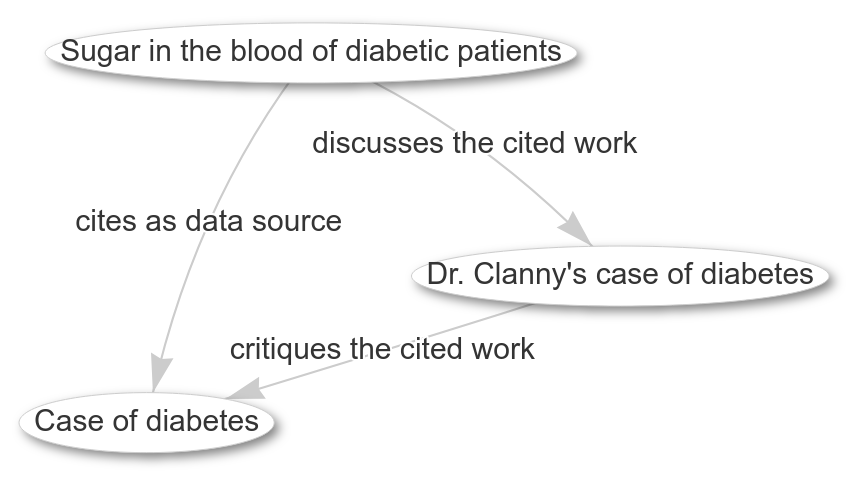
This network is based on public knowledge in Wikidata and actually can be easily reproduced by anyone with this query:
#defaultView:Graph
SELECT DISTINCT ?focus1 ?focus1Label ?focus2 ?focus2Label ?edgeLabel WHERE {
VALUES ?focus1 { wd:Q124174475 wd:Q124174776 wd:Q124174815 }
VALUES ?focus2 { wd:Q124174475 wd:Q124174776 wd:Q124174815 }
?focus1 p:P2860 ?citation .
?citation ps:P2860 ?focus2 ; pq:P3712 ?edge .
?edge rdfs:label ?edgeLabel . FILTER(LANG(?edgeLabel) = "en")
SERVICE wikibase:label { bd:serviceParam wikibase:language "[AUTO_LANGUAGE],mul,en". }
}
The two “focus” values are an identical list of the articles I want to see. To make sure to get citations between all of them, I have to give them twice.
In the above example I have used VALUES for this, but I can also generate the controlled list of items between the citations
I want to visualize with any SPARQL fragment too. This query does that (or here as
GitHub Gist, but something else too: it uses a trick I learned
from Finn Nielsen from this patch
from the Scholia project (doi:10.1007/978-3-319-70407-4_36)).
Here, I select the articles by replacing the above VALUES lines with this fragment (P50 is ‘author’ and Q20895241 is me in Wikidata):
?focus1 wdt:P50 wd:Q20895241 .
?focus2 wdt:P50 wd:Q20895241 .
And, to be honest, then I get this network which is much richer than I expected:

I wonder how far we can push this. Can we also do this for the Journal of Cheminformatics? After all, this journal had a CiTO Pilot and, indeed, the results do not disappoint! All I had to do was replace the focus section:
?focus1 wdt:P1433 wd:Q6294930 .
?focus2 wdt:P1433 wd:Q6294930 .