New paper: "Discovering life's directed metabolic (sub)paths to interpret human biochemical markers using the DSMN tool"
I am still catching up with a lot of work, and found out I actually had forgotten to blog about this cool article by Denise Slenter: “Discovering life’s directed metabolic (sub)paths to interpret human biochemical markers using the DSMN tool” (doi:10.1039/D3DD00069A). This paper explains how various open science resources (Wikidata, Reactome, WikiPathways) are used to visualize the biological story of the data from two metabolomics experiments archived in MetaboLights.
Using Neo4J and Cytoscape she visualizes the data onto a network created with RDF, SPARQL from the above resources:
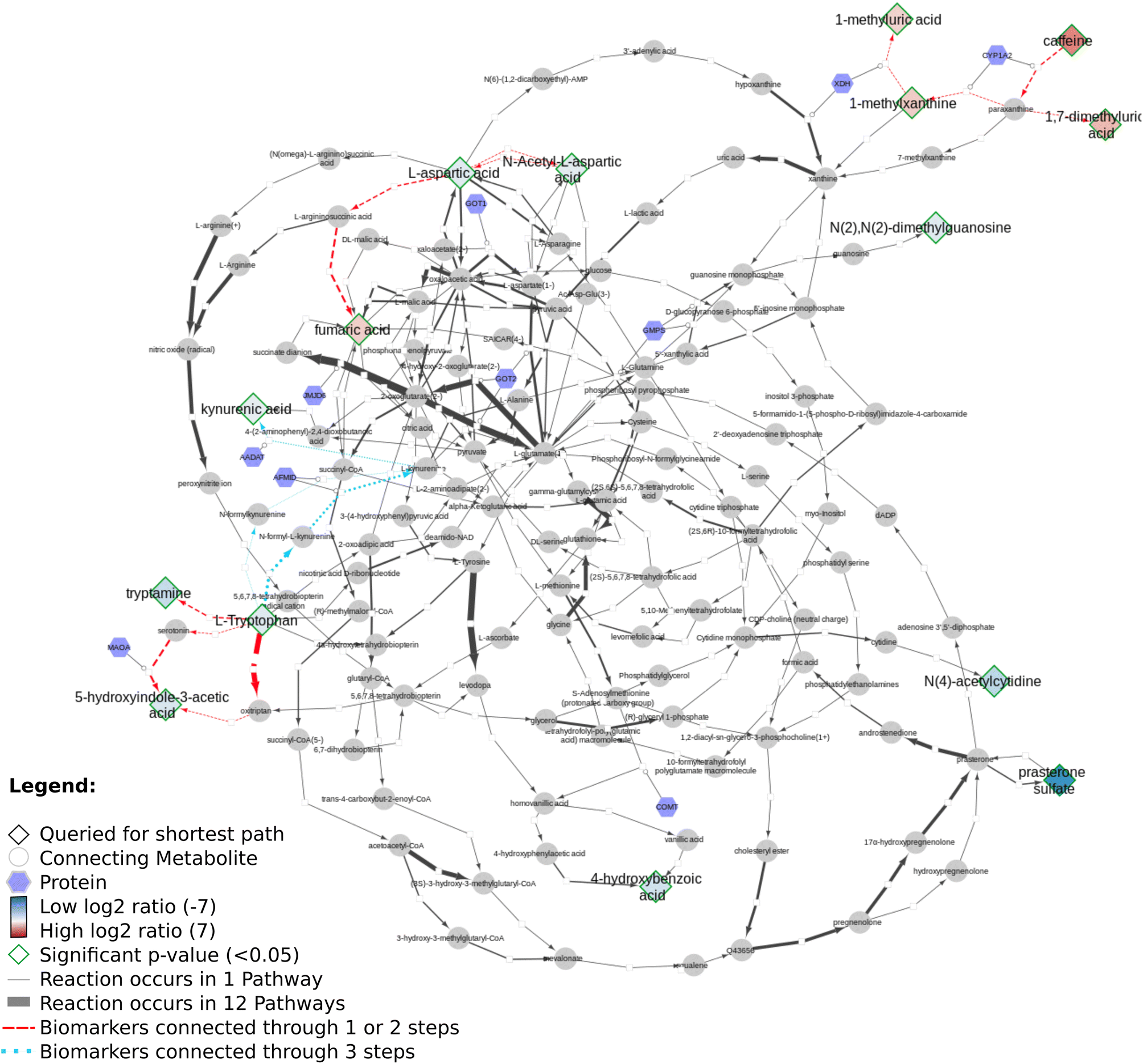
The whole approach uses open science, making the work very reproducible. This is essential, as our knowledge about metabolic processes continues to grow, if not only for the human lipids, but also from molecular imaging technologies. Moreover, a lot of biological detail is yet to be encoded on pathway databases, such as cellular location of proteins and metabolites, which proteins are expressed in which tissue, or the kinetics of metabolic reactions. All knowledge that can be pulled it via knowledge graphs becomes immediately available by using this FAIR approach.
One last note, the reader may notice a focus on the shortest path. Of course, the biological relevant path may not be the “shortest” path. But from a network analysis perspective that question is purely academic. Neo4J, like other tools, support finding all paths. But validation which paths (the shorter or any of the longer) is biologically most relevant first depends on actually more biological knowledge to become FAIR. After this, it is just push button.