new: "Providing Adverse Outcome Pathways from the AOP-Wiki in a Semantic Web Format to Increase Usability and Accessibility of the Content"
I am a bit behind with tweeting about new published papers, but let that not reflect that these papers are not very exciting. The first paper is by Marvin an almost-finished PhD candidate in our group and now working as postdoc on the VHP4Safety project. He has been working on linking adverse outcome pathways (AOPs) with molecular pathways, such as in WikiPathways. This work was mostly done as part of the EU projects OpenRiskNet and EUToxRisk , during which he disseminated his research in many directions (e.g. the second paper in this post). Talking about impact.
He previously already sketched out the ideas of integration the two kinds of pathways in this paper. The implementation of this has now been published in the paper Providing Adverse Outcome Pathways from the AOP-Wiki in a Semantic Web Format to Increase Usability and Accessibility of the Content (doi:10.1089/aivt.2021.0010). It’s an important piece of our growing molecular life sciences knowledge graph, which already contains data from WikiPathways and ChEMBL. Of course, integrated with other SPARQL endpoints, such as NextProt/UniProt, Rhea, etc.
Schematic diagram from the article showing the kind of information in the database:
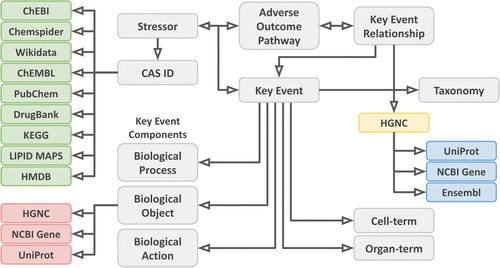
Marvin writes: “The resulting RDF contains >122,000 triples describing 158 unique properties of >15,000 unique subjects. Furthermore, >3500 link-outs were added to 12 chemical databases, and >7500 link-outs to 4 gene and protein databases. The AOP-Wiki RDF has been made available at https://aopwiki.rdf.bigcat-bioinformatics.org”. The last comes with many example queries.