SARS-CoV-2, COVID-19, and Open Science
Voices are getting stronger over how important Open Science is. Insiders have known the advantages for decades. We also know the issues in the transition, but the transition has been steady. Contributing to Open Science is simple: there are plenty of project where you can contribute without jeopardizing your own research (funding or prestige). Myself, my small contributions have been done without funding too. But I needed to do something. I have been mostly self-quarantined since March 6, with only very few exception. And I’m so done with it. Like so many other people. It won’t stop me wearing masks when I go shopping (etc).
Reflecting on the past eight months, particularly the last two months have been tough. It’s easier to sit at home and in the garden when it is warm outside, light. But for another 7 weeks or so, they days will only get darker. The past two months were also so busy with grant reporting that I did not get around to much else, even with an uncommon long stretch of long working weeks, with about 8 weeks of 70-80 hrs of active work in that period. In fact, the past two weeks, with most of the deadlines past, I had a physical reset, and was happy if I made 40 hrs a week.
So, where is my COVID-19 work now, where is it going?
Molecular Pathways
First, what did we reach? First, leveraging from the Open Science community I am involved in, I stared collaborating. With old friends and making new friends. I was delighted to see I was not the only one. In fact, Somewhere in May/June, I had to give up following all Open Science around COVID-19, because there was too much.
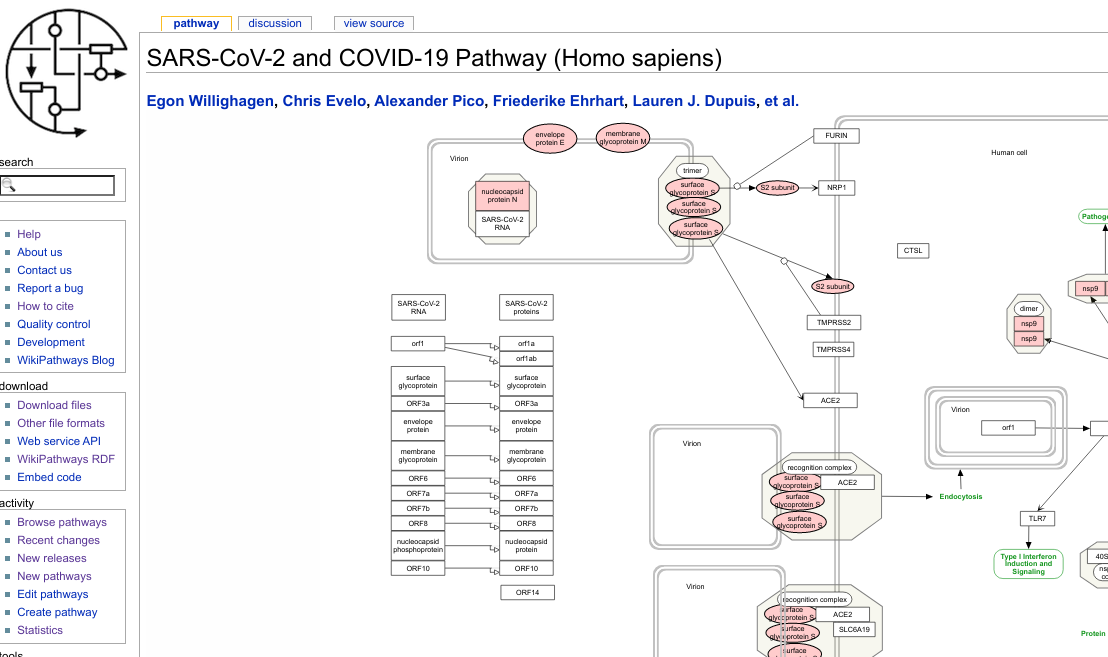
For example, I was not the only one wanting to describe our slowly developing molecular knowledge of the SARS-CoV-2 virus. While my pathway focused on specifically the confirmed processes for SARS-CoV-2, my colleague Freddie digitized a recent review about other corona viruses. Check out her work: WP4863, WP4864, WP4877, WP4880, and WP4912. In fact, In fact, so much was done by so many people in such a short time, that the WikiPathways COVID-19 Portal was set up.
Further reading:
- Ostaszewski M. COVID-19 Disease Map, a computational knowledge repository of SARS-CoV-2 virus-host interaction mechanisms. bioRxiv. 2020 Oct 28; 10.1101/2020.10.26.356014v1 (and unversioned 10.1101/2020.10.26.356014)
- Ostaszewski M, Mazein A, Gillespie ME, Kuperstein I, Niarakis A, Hermjakob H, et al. COVID-19 Disease Map, building a computational repository of SARS-CoV-2 virus-host interaction mechanisms. Sci Data. 2020 May 5;7(1):136 10.1038/s41597-020-0477-8
Interoperability with Wikidata
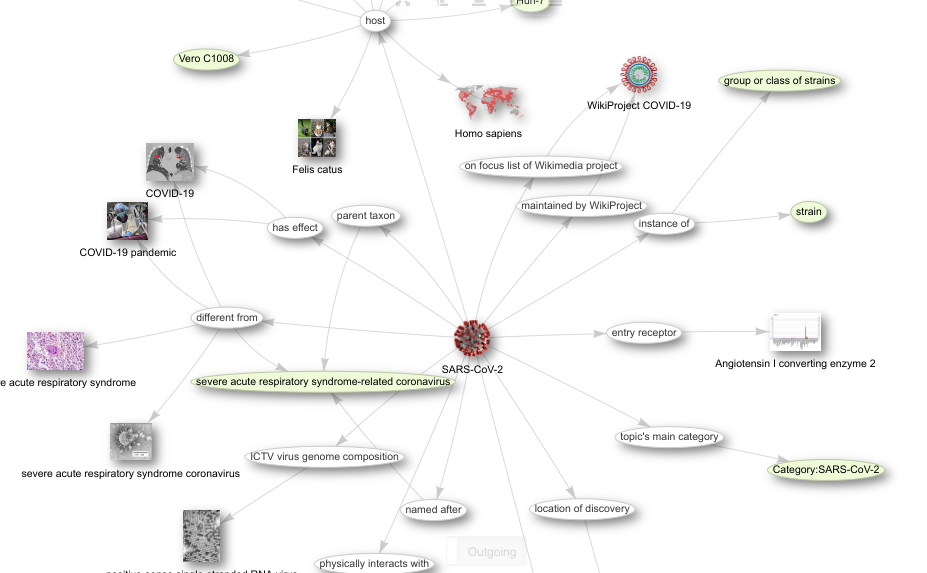
Because I see an essential role for Wikidata in Open Science, and because regular databases did not provide identifiers for the molecular building blocks, we created them in Wikidata. This was essential, because I wanted to use Scholia (see screenshot on the right) to track the research output (something that by now has become quite a challenge; btw, checkout Lauren’s tutorial on this). This too was still in March. However, because Scholia itself is a general tool, I needed shortlists of all SARS-CoV-2 genes, all proteins, etc. So, I created this book. It’s autogenerated and auto-updated by taking advantage of SPARQL queries against Wikidata. And I am so excited the book has been translated in Japanese, Portugues, and Spanish. The i18n work is thanks to the virtual BioHackathon in April, where Yayamamo bootstrapped the framework to localize the content.
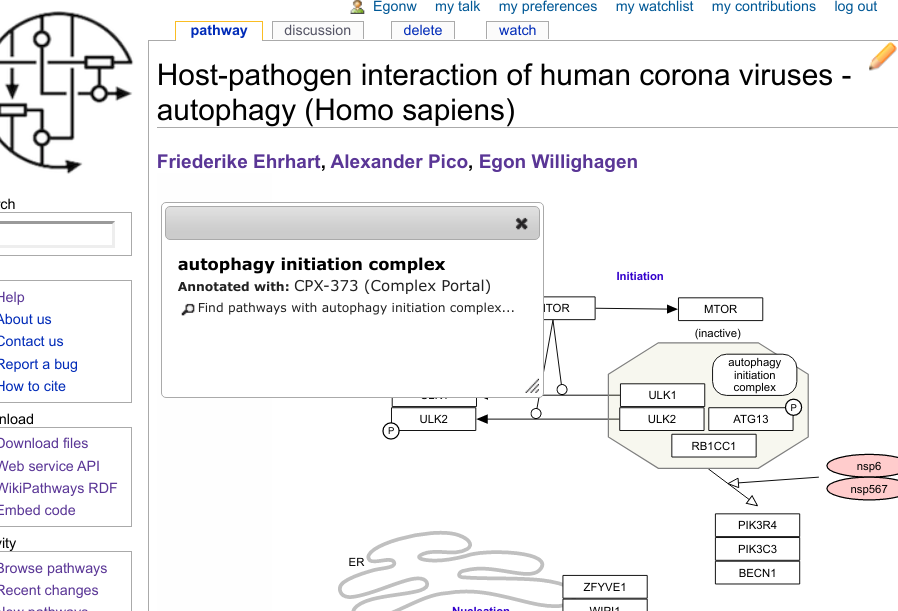
Also during that BioHackathon, we started a collaboration with Complex Portal’s Birgit, because the next step was to have identifiers for (bio)molecular complexes. This work is still ongoing, but using a workaround we developed for WikiPathways (because complexes in GPML currently cannot have identifiers), we can now link out to Complex Portal, as visible in this screenshot:
Joining the Wikidata effort is simple. Just visit Wikidata:WikiProject_COVID-19 and find your thing of interest. Because the past two months have been so crowded, I still did not get around to explore the kg-covid-19 project, but sounds very interesting too!
Further reading:
- Waagmeester A, Stupp G, Burgstaller-Muehlbacher S, Good BM, Griffith M, Griffith OL, et al. Wikidata as a knowledge graph for the life sciences. eLife. 2020 Mar 17;9:e52614. 10.7554/eLife.52614
- Waagmeester A, Willighagen EL, Su AI, Kutmon M, Labra Gayo JE, Fernández-Álvarez D, et al. A protocol for adding knowledge to Wikidata, a case report. bioRxiv [Internet]. 2020 Apr 7 [cited 2020 Apr 17]; 10.1101/2020.04.05.026336
Computer-assisted data curation
For some years now, I have been working on computer-assisted data curation of WikiPathways, but also Wikidata (for chemical compounds). Once your biological knowledge is machine readable, you can have learn machines to recognize common mistakes. Some are basically simple checks, like missing information. But it gets exciting if we take advantage of linked data, and we can have machines check consistency between two or more resources. The better out annotation, the more powerful this computer-assisted data curation becomes. Chris has been urging me to publish this, but I haven’t gotten around to this yet.
As part of my COVID-19 work, I have started making curation reports for specific WikiPathways. To enable this, I worked out how to reuse the testing without JUnit, allowing the tests to be used as a library. That allows creating the reports, but in the future will also allow use directly in PathVisio. A second improvement to the testing stack is that tests are now more easily annotated. That allows specifying tests only to be run for a certain WikiPathways portal.
But a lot remains to be done. I think at this moment I only migrated, perhaps, some 5% of all tests. So, this is very much on my “what is next?” list.
What is next?
There is a lot I need, want, and should do. here are some ideas. Maybe you wan to beat me to it. Really, I don’t mind being scooped, when it comes to public health. Here goes:
- file SARS-CoV-2 book translation update requests for some recent updates
- update the SARS-CoV-2 book with a list of important SNPs
- add BioSchemase to the SARS-CoV-2 book for individual proteins, genes, etc
- update WP4846 with recent literature
- have another ‘main subject’ annotation round for SARS-CoV-2 proteins
- migrate more pathways tests from JUnit into the testing library
- write a new test to detect preprints in pathway literature lists and check for journal article versions
- finish the Dutch translation of the SARS-CoV-2 book
- write a tool to recognize WikiPathways complexes with matches in Complex Portal
- write a tool to generate markdown for any WikiPathways with curation suggestions based on content in other resources
- develop a few HTML+JavaScript pages to summarize WikiPathways COVID-19 Portal content
Am I missing anything? Tweet me or leave a comment here.