Nature Chemistry improves publishing chemistry: a detailed analysis
Nature Chemistry just released the first issue with a few free papers, like Asymmetric total syntheses of (+)- and (-)-versicolamide B and biosynthetic implications by Miller et al. (DOI:10.1038/nchem.110).
Now, we’ve seen the Royal Society of Chemistry’s Project Prospect (see RSC: the first publisher to go semantic! ) and ChemSpiders recent ChemMantis system which enriches the papers with machine readable representations of the molecules discussed in those papers. The new Nature publication has been in the works for a while, and they asked the community before what a Nature Chemistry paper should like like, and I replied in Re: What should a Nature Chemistry paper look like? .
The verdict
So, have the been listening? Is the HTML they produce semantic? Is it data rich? Or is it just another hamburger? Well, I am very happy to see some of the suggestions I made picked up (though I do not fool myself in believing I am the only one that suggested those features). A tour of good things, and points for improvement.
The first impression is not shocking; it looks like any other interface, with molecules drawn as images in the paper:
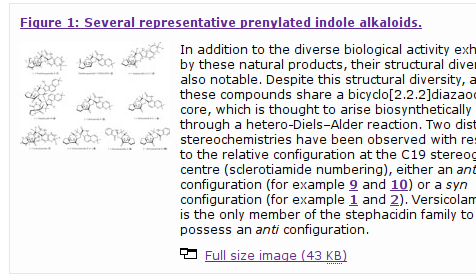
All structures that are numbered and linked (as in C6-epi-stephacidin A (Compound 13) have a hover-over function to popup a drawing of the structure:
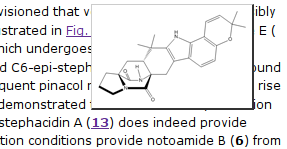
The popup image is a nice gimmick, but not really sematically useful. The link, however, is! It points to a separate supplementary page with further information which include a image of the 2D structure and, following a link, the 3D structure in Jmol. Moreover, it comes with the machine readable representations:

This is indeed interesting, and a big step forward, though please do note my comments later. For convenience, all molecules with such supplementary information is available from the special Chemical Compounds section of the paper:

Excellent! This really is a step forward towards a data-rich paper! Indeed, I will shortly write up a Bioclipse plugin for Nature Chemistry, which will download all molecular structures based on the DOI! Anyway, more on that later… For this article, that table looks like:

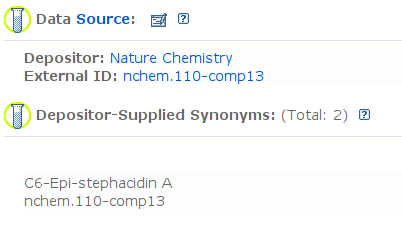
By now, you likely also noted the links to PubChem, and indeed, upon publication of a paper, all structures are deposited in the public domain:
At last but not least, each molecule is available in the Chemical Markup Language (with 2D coordinates)! And you know I am a very happy CML user for a long time (see e.g. Peter’s recent blog Egon Willighagen and CML ). BTW, one comment on the CML: the namespace used is the outdated namespace, not the current one (see There can be only one (namespace)). (But the CDK and Bioclipse will read it anyway.)
Details matter
So, while the first impression was not shocking, it was a bit deceptive. Nature Chemistry really changes publishing of chemistry. But I have bad news too. They need to improve the HTML they produce.
But before pointing out some missed chances, let me reply inter alia to Peter’s recent work on the Open Source plugin for including semantic chemistry in MS-Word documents (see [How can we publish semantic chemical documents? </i](https://blogs.ch.cam.ac.uk/pmr/2009/03/16/how-can-we-publish-semantic-chemical-documents/)): Nature Chemistry seems to have done a great job with existing tools. Nevertheless, I fully back up Peters comment that while the plugin is useless without Word, the results produced with the plugin are extremely Open Standard, and enormously reusable! Indeed, while the Word file format is only formally an true Open Standard, the file format is plain XML, and extracting content bearing the CML namespace is trivial.
Which reminds me, if someone from the Nature Chemistry team is reading this, please point me to a blog what tools actually are involved in publishing a Nature Chemistry paper! I think we all like to know.
Now, the HTML has room for improvement. First of all, a look at the metadata defined for the web page of the article shows a description and keywords about the journal, not the article, and the same goes for the web pages for the molecules:

Additionally, the compound details web page has no special markup for the machine readable information:
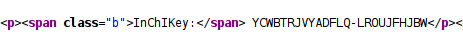
Or, if it does, it’s still mixed with markup for visual pleasing output:

Still, the HTML is clean enough to have some regular expressions extract a good deal of information, and there is also still the PubChem deposition.
Beyond connection tables
Like many other chemistry journals, Nature Chemistry does not consider properties of the molecule interesting, and NMR spectra are hidden in the Supplementary Information. This paper in particular, disregards a lot of machine readable facts by putting all experimental section bits in a PDF document. So, the next challenge for Nature Chemistry will be to get the authors of papers contribute the original spectra (JCAMP-DX, CMLSpect, etc) in the supplementary information section. Better, have the raw data or even the NMR peak-atom annotations deposited in public repositories such (see Open NMR data: raw curves and annotated peak lists ).
All in all, I am rather positive about the first Nature Chemistry issue, and like to thank the editors and paper authors for there efforts on improving publishing chemistry!